High-throughput mutagenesis using a two-fragment PCR approach
- PMID: 28754896
- PMCID: PMC5533798
- DOI: 10.1038/s41598-017-07010-4
High-throughput mutagenesis using a two-fragment PCR approach
Abstract
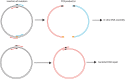
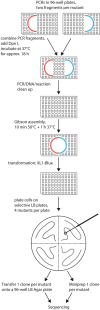
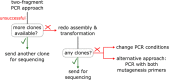
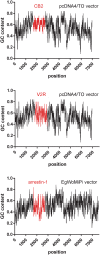
Site-directed scanning mutagenesis is a powerful protein engineering technique which allows studies of protein functionality at single amino acid resolution and design of stabilized proteins for structural and biophysical work. However, creating libraries of hundreds of mutants remains a challenging, expensive and time-consuming process. The efficiency of the mutagenesis step is the key for fast and economical generation of such libraries. PCR artefacts such as misannealing and tandem primer repeats are often observed in mutagenesis cloning and reduce the efficiency of mutagenesis. Here we present a high-throughput mutagenesis pipeline based on established methods that significantly reduces PCR artefacts. We combined a two-fragment PCR approach, in which mutagenesis primers are used in two separate PCR reactions, with an in vitro assembly of resulting fragments. We show that this approach, despite being more laborious, is a very efficient pipeline for the creation of large libraries of mutants.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cunningham, B. C. & Wells, J. A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science244, 1081–1085, doi:10.1126/science.2471267 (1989). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

