Epigenetic changes in myelofibrosis: Distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations
- PMID: 28754985
- PMCID: PMC5533802
- DOI: 10.1038/s41598-017-07057-3
Epigenetic changes in myelofibrosis: Distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations
Erratum in
-
Publisher Correction: Epigenetic changes in myelofibrosis: Distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations.Sci Rep. 2018 Nov 20;8(1):17311. doi: 10.1038/s41598-018-35596-w. Sci Rep. 2018. PMID: 30459458 Free PMC article.
Abstract
This is the first study to compare genome-wide DNA methylation profiles of sorted blood cells from myelofibrosis (MF) patients and healthy controls. We found that differentially methylated CpG sites located to genes involved in 'cancer' and 'embryonic development' in MF CD34+ cells, in 'inflammatory disease' in MF mononuclear cells, and in 'immunological diseases' in MF granulocytes. Only few differentially methylated CpG sites were common among the three cell populations. Mutations in the epigenetic regulators ASXL1 (47%) and TET2 (20%) were not associated with a specific DNA methylation pattern using an unsupervised approach. However, in a supervised analysis of ASXL1 mutated versus wild-type cases, differentially methylated CpG sites were enriched in regions marked by histone H3K4me1, histone H3K27me3, and the bivalent histone mark H3K27me3 + H3K4me3 in human CD34+ cells. Hypermethylation of selected CpG sites was confirmed in a separate validation cohort of 30 MF patients by pyrosequencing. Altogether, we show that individual MF cell populations have distinct differentially methylated genes relative to their normal counterparts, which likely contribute to the phenotypic characteristics of MF. Furthermore, differentially methylated CpG sites in ASXL1 mutated MF cases are found in regulatory regions that could be associated with aberrant gene expression of ASXL1 target genes.
Conflict of interest statement
VP is consultant in bioinfreg.
Figures

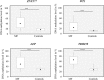
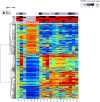
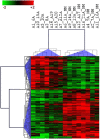

References
-
- Barosi G. Myelofibrosis with myeloid metaplasia: diagnostic definition and prognostic classification for clinical studies and treatment guidelines. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1999;17:2954–2970. doi: 10.1200/JCO.1999.17.9.2954. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

