Zinc binding to RNA recognition motif of TDP-43 induces the formation of amyloid-like aggregates
- PMID: 28754988
- PMCID: PMC5533730
- DOI: 10.1038/s41598-017-07215-7
Zinc binding to RNA recognition motif of TDP-43 induces the formation of amyloid-like aggregates
Abstract
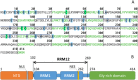
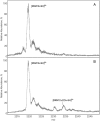
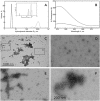
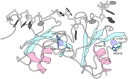
Aggregation of TDP-43 (transactive response DNA binding protein 43 kDa) is a hallmark of certain forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Moreover, intracellular TDP-43-positive inclusions are often found in other neurodegenerative diseases. Recently it was shown that zinc ions can provoke the aggregation of endogenous TDP-43 in cells, allowing to assume a direct interaction of TDP-43 with zinc ions. In this work, we investigated zinc binding to the 102-269 TDP-43 fragment, which comprise the two RNA recognition motifs. Using isothermal titration calorimetry, mass spectrometry, and differential scanning fluorimetry, we showed that zinc binds to this TDP-43 domain with a dissociation constant in the micromolar range and modifies its tertiary structure leading to a decrease of its thermostability. Moreover, the study by dynamic light scattering and negative stain electron microscopy demonstrated that zinc ions induce auto-association process of this TDP-43 fragment into rope-like structures. These structures are thioflavin-T-positive allowing to hypothesize the direct implication of zinc ions in pathological aggregation of TDP-43.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Buratti E. Functional Significance of TDP-43 Mutations in Disease. Adv Genet. 2015;91:1–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

