A novel family of proline/serine-rich proteins, which are phospho-targets of stress-related mitogen-activated protein kinases, differentially regulates growth and pathogen defense in Arabidopsis thaliana
- PMID: 28755319
- PMCID: PMC5594048
- DOI: 10.1007/s11103-017-0641-5
A novel family of proline/serine-rich proteins, which are phospho-targets of stress-related mitogen-activated protein kinases, differentially regulates growth and pathogen defense in Arabidopsis thaliana
Abstract
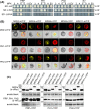
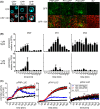
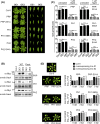
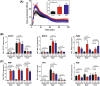
The molecular actions of mitogen-activated protein kinases (MAPKs) are ultimately accomplished by the substrate proteins where phosphorylation affects their molecular properties and function(s), but knowledge regarding plant MAPK substrates is currently still fragmentary. Here, we uncovered a previously uncharacterized protein family consisting of three proline/serine-rich proteins (PRPs) that are substrates of stress-related MAPKs. We demonstrated the importance of a MAPK docking domain necessary for protein-protein interaction with MAPKs and consequently also for phosphorylation. The main phosphorylated site was mapped to a residue conserved between all three proteins, which when mutated to a non-phosphorylatable form, differentially affected their protein stability. Together with their distinct gene expression patterns, this differential accumulation of the three proteins upon phosphorylation probably contributes to their distinct function(s). Transgenic over-expression of PRP, the founding member, led to plants with enhanced resistance to Pseudomonas syringae pv. tomato DC3000. Older plants of the over-expressing lines have curly leaves and were generally smaller in stature. This growth phenotype was lost in plants expressing the phosphosite variant, suggesting a phosphorylation-dependent effect. Thus, this novel family of PRPs may be involved in MAPK regulation of plant development and / or pathogen resistance responses. As datamining associates PRP expression profiles with hypoxia or oxidative stress and PRP-overexpressing plants have elevated levels of reactive oxygen species, PRP may connect MAPK and oxidative stress signaling.
Keywords: MAPK; Oxidative stress; PAMPs; Pathogen resistance; Phosphorylation; Plant development.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

