Synthetic lethal short hairpin RNA screening reveals that ring finger protein 183 confers resistance to trametinib in colorectal cancer cells
- PMID: 28756770
- PMCID: PMC5535279
- DOI: 10.1186/s40880-017-0228-1
Synthetic lethal short hairpin RNA screening reveals that ring finger protein 183 confers resistance to trametinib in colorectal cancer cells
Abstract
Background: The mitogen-activated extracellular signal-regulated kinase 1/2 (MEK1/2) inhibitor trametinib has shown promising therapeutic effects on melanoma, but its efficacy on colorectal cancer (CRC) is limited. Synthetic lethality arises with a combination of two or more separate gene mutations that causes cell death, whereas individual mutations keep cells alive. This study aimed to identify the genes responsible for resistance to trametinib in CRC cells, using a synthetic lethal short hairpin RNA (shRNA) screening approach.
Methods: We infected HT29 cells with a pooled lentiviral shRNA library and applied next-generation sequencing to identify shRNAs with reduced abundance after 8-day treatment of 20 nmol/L trametinib. HCT116 and HT29 cells were used in validation studies. Stable ring finger protein 183 (RNF183)-overexpressing cell lines were generated by pcDNA4-myc/his-RNF183 transfection. Stable RNF183-knockdown cell lines were generated by infection of lentiviruses that express RNF183 shRNA, and small interference RNA (siRNA) was used to knock down RNF183 transiently. Quantitative real-time PCR was used to determine the mRNA expression. Western blotting, immunohistochemical analysis, and enzyme-linked immunosorbent assay (ELISA) were used to evaluate the protein abundance. MTT assay, colony formation assay, and subcutaneous xenograft tumor growth model were used to evaluate cell proliferation.
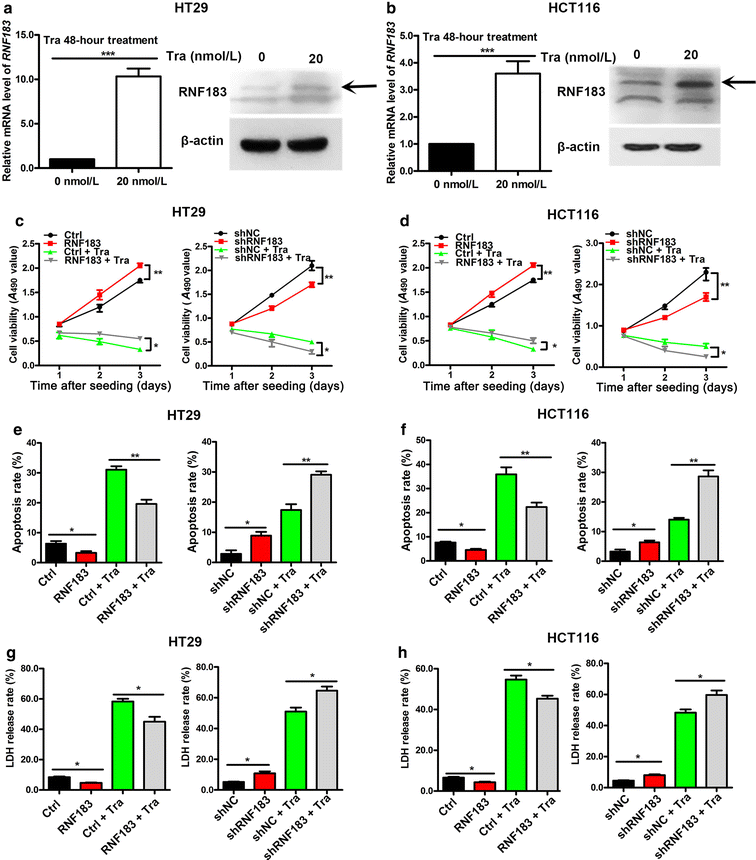
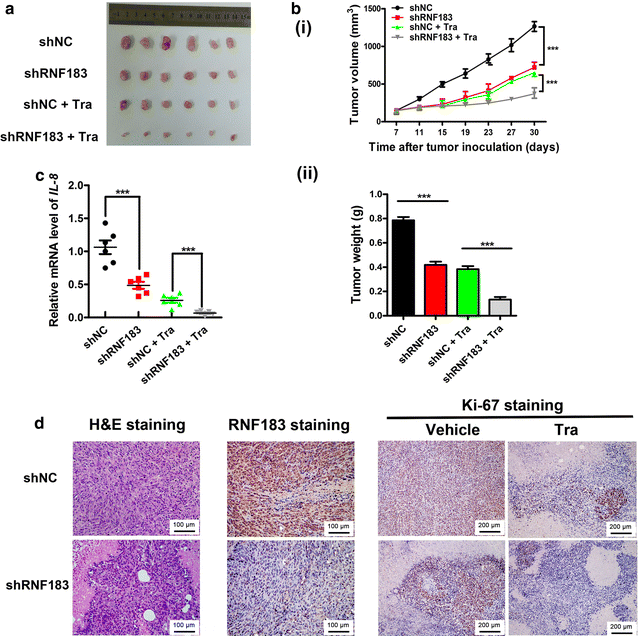
Results: In the primary screening, we found that the abundance of RNF183 shRNA was markedly reduced after treatment with trametinib. Trametinib induced the expression of RNF183, which conferred resistance to drug-induced cell growth repression and apoptotic and non-apoptotic cell deaths. Moreover, interleukin-8 (IL-8) was a downstream gene of RNF183 and was required for the function of RNF183 in facilitating cell growth. Additionally, elevated RNF183 expression partly reduced the inhibitory effect of trametinib on IL-8 expression. Finally, xenograft tumor model showed the synergism of RNF183 knockdown and trametinib in repressing the growth of CRC cells in vivo.
Conclusion: The RNF183-IL-8 axis is responsible for the resistance of CRC cells to the MEK1/2 inhibitor trametinib and may serve as a candidate target for combined therapy for CRC.
Keywords: Colorectal cancer; Mitogen-activated extracellular signal-regulated kinase 1/2; Ring finger protein 183; Synthetic lethal; Trametinib.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

