The house spider genome reveals an ancient whole-genome duplication during arachnid evolution
- PMID: 28756775
- PMCID: PMC5535294
- DOI: 10.1186/s12915-017-0399-x
The house spider genome reveals an ancient whole-genome duplication during arachnid evolution
Abstract
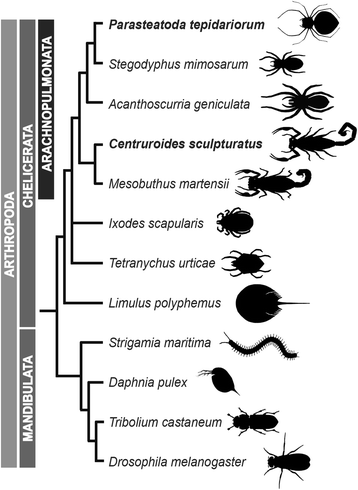
Background: The duplication of genes can occur through various mechanisms and is thought to make a major contribution to the evolutionary diversification of organisms. There is increasing evidence for a large-scale duplication of genes in some chelicerate lineages including two rounds of whole genome duplication (WGD) in horseshoe crabs. To investigate this further, we sequenced and analyzed the genome of the common house spider Parasteatoda tepidariorum.
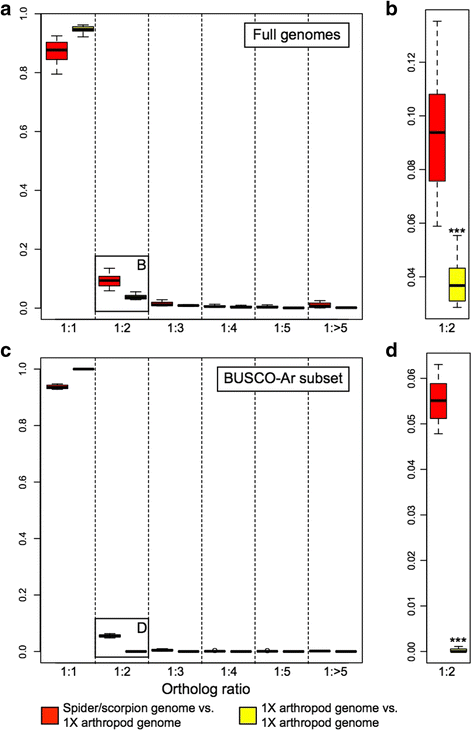
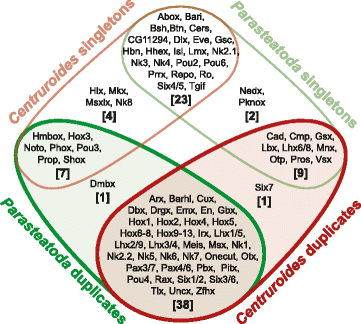
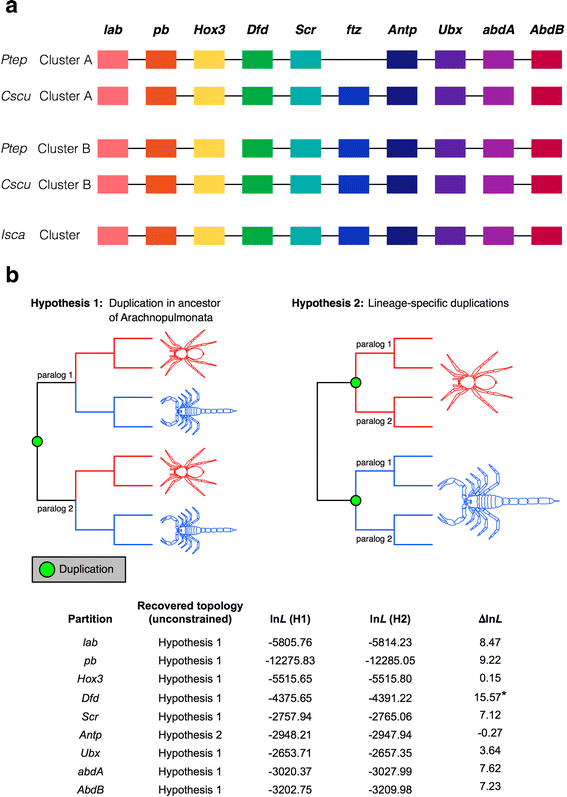
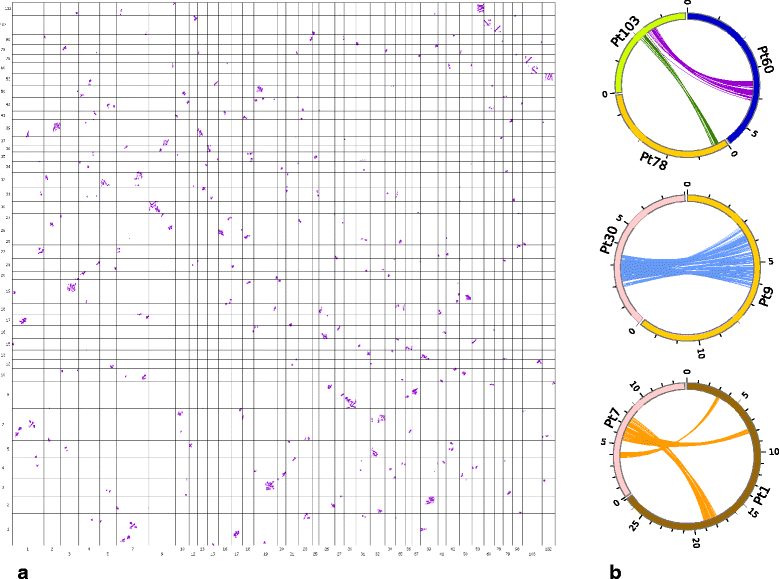
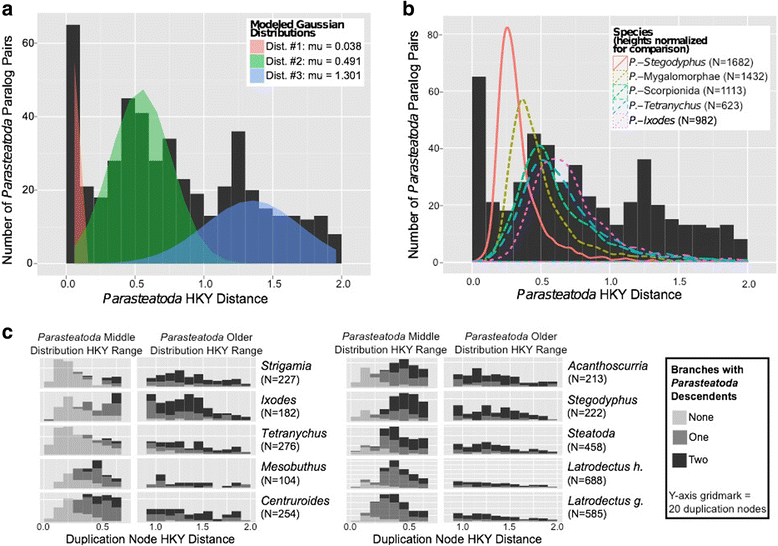
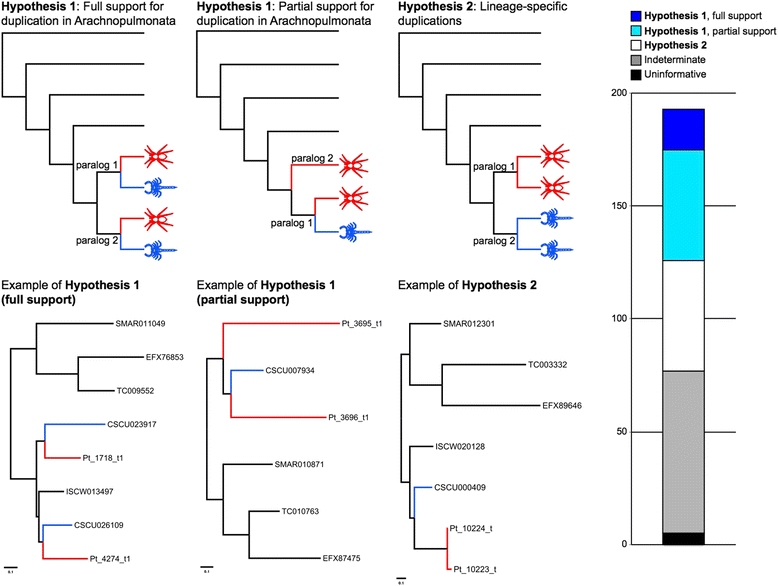
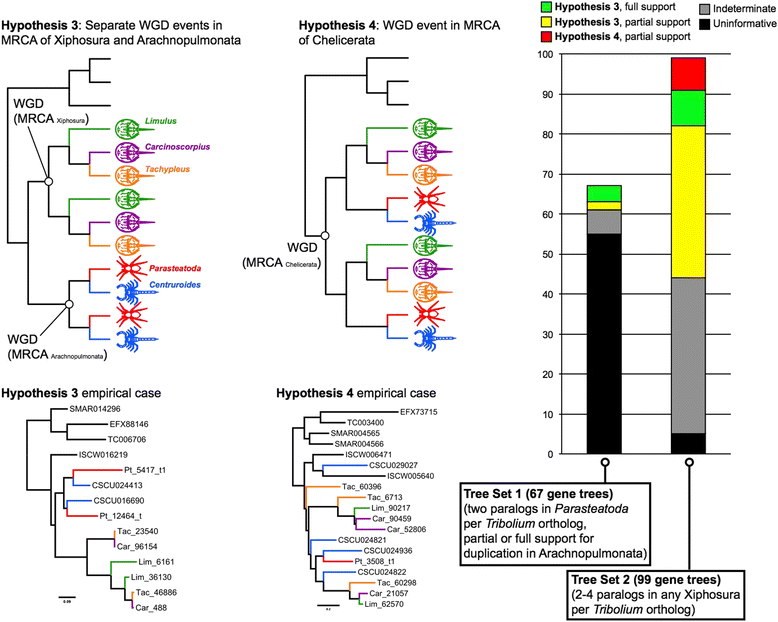
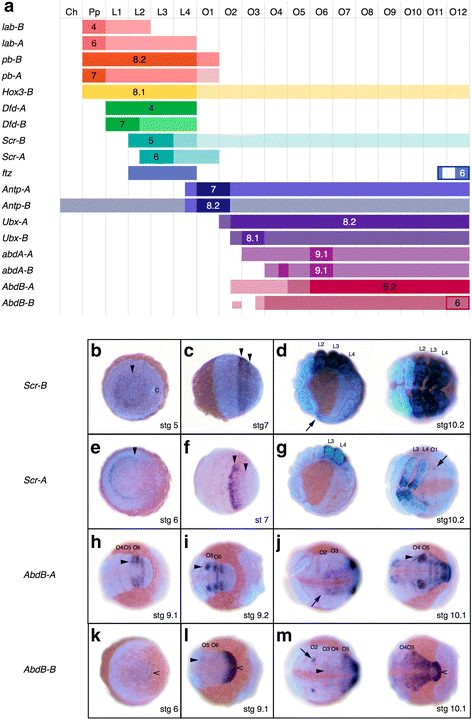
Results: We found pervasive duplication of both coding and non-coding genes in this spider, including two clusters of Hox genes. Analysis of synteny conservation across the P. tepidariorum genome suggests that there has been an ancient WGD in spiders. Comparison with the genomes of other chelicerates, including that of the newly sequenced bark scorpion Centruroides sculpturatus, suggests that this event occurred in the common ancestor of spiders and scorpions, and is probably independent of the WGDs in horseshoe crabs. Furthermore, characterization of the sequence and expression of the Hox paralogs in P. tepidariorum suggests that many have been subject to neo-functionalization and/or sub-functionalization since their duplication.
Conclusions: Our results reveal that spiders and scorpions are likely the descendants of a polyploid ancestor that lived more than 450 MYA. Given the extensive morphological diversity and ecological adaptations found among these animals, rivaling those of vertebrates, our study of the ancient WGD event in Arachnopulmonata provides a new comparative platform to explore common and divergent evolutionary outcomes of polyploidization events across eukaryotes.
Keywords: Centruroides sculpturatus; Evolution; Gene duplication; Genome; Hox genes; Parasteatoda tepidariorum.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970.
-
- Chipman AD, Ferrier DE, Brena C, Qu J, Hughes DS, Schroder R, Torres-Oliva M, Znassi N, Jiang H, Almeida FC, et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12(11) doi: 10.1371/journal.pbio.1002005. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

