Vps34 Kinase Domain Dynamics Regulate the Autophagic PI 3-Kinase Complex
- PMID: 28757208
- PMCID: PMC5573195
- DOI: 10.1016/j.molcel.2017.07.003
Vps34 Kinase Domain Dynamics Regulate the Autophagic PI 3-Kinase Complex
Abstract
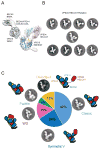
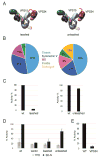
The class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1) is required for the initiation of essentially all macroautophagic processes. PI3KC3-C1 consists of the lipid kinase catalytic subunit VPS34, the VPS15 scaffold, and the regulatory BECN1 and ATG14 subunits. The VPS34 catalytic domain and BECN1:ATG14 subcomplex do not touch, and it is unclear how allosteric signals are transmitted to VPS34. We used EM and crosslinking mass spectrometry to dissect five conformational substates of the complex, including one in which the VPS34 catalytic domain is dislodged from the complex but remains tethered by an intrinsically disordered linker. A "leashed" construct prevented dislodging without interfering with the other conformations, blocked enzyme activity in vitro, and blocked autophagy induction in yeast cells. This pinpoints the dislodging and tethering of the VPS34 catalytic domain, and its regulation by VPS15, as a master allosteric switch in autophagy induction.
Keywords: allostery; autophagy; crosslinking mass spectrometry; electron microscopy; lipid kinase; phosphoinositide; protein dynamics; protein kinase; protein structure; pseudokinase.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

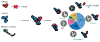
References
-
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. - PubMed
-
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. - PubMed
-
- Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J. 2016;473:2251–2271. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

