Beyond mRNA: The role of non-coding RNAs in normal and aberrant hematopoiesis
- PMID: 28757239
- PMCID: PMC5722683
- DOI: 10.1016/j.ymgme.2017.07.008
Beyond mRNA: The role of non-coding RNAs in normal and aberrant hematopoiesis
Abstract
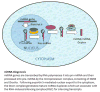
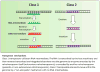
The role of non-coding Ribonucleic Acids (ncRNAs) in biology is currently an area of intense focus. Hematopoiesis requires rapidly changing regulatory molecules to guide appropriate differentiation and ncRNA are well suited for this. It is not surprising that virtually all aspects of hematopoiesis have roles for ncRNAs assigned to them and doubtlessly much more await characterization. Stem cell maintenance, lymphoid, myeloid and erythroid differentiation are all regulated by various ncRNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and various transposable elements within the genome. As our understanding of the many and complex ncRNA roles continues to grow, new discoveries are challenging the existing classification schemes. In this review we briefly overview the broad categories of ncRNAs and discuss a few examples regulating normal and aberrant hematopoiesis.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


References
-
- Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nature structural & molecular biology. 2015;22(1):11–9. Epub 2015/01/08. - PubMed
-
- Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nature reviews Genetics. 2014;15(3):193–204. Epub 2014/02/12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

