Precision Functional Mapping of Individual Human Brains
- PMID: 28757305
- PMCID: PMC5576360
- DOI: 10.1016/j.neuron.2017.07.011
Precision Functional Mapping of Individual Human Brains
Abstract
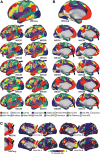
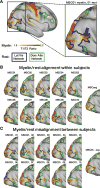
Human functional MRI (fMRI) research primarily focuses on analyzing data averaged across groups, which limits the detail, specificity, and clinical utility of fMRI resting-state functional connectivity (RSFC) and task-activation maps. To push our understanding of functional brain organization to the level of individual humans, we assembled a novel MRI dataset containing 5 hr of RSFC data, 6 hr of task fMRI, multiple structural MRIs, and neuropsychological tests from each of ten adults. Using these data, we generated ten high-fidelity, individual-specific functional connectomes. This individual-connectome approach revealed several new types of spatial and organizational variability in brain networks, including unique network features and topologies that corresponded with structural and task-derived brain features. We are releasing this highly sampled, individual-focused dataset as a resource for neuroscientists, and we propose precision individual connectomics as a model for future work examining the organization of healthy and diseased individual human brains.
Keywords: brain networks; fMRI; functional connectivity; individual variability; myelin mapping.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

