Different Metabolic Pathways Are Involved in Response of Saccharomyces cerevisiae to L-A and M Viruses
- PMID: 28757599
- PMCID: PMC5577567
- DOI: 10.3390/toxins9080233
Different Metabolic Pathways Are Involved in Response of Saccharomyces cerevisiae to L-A and M Viruses
Abstract
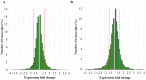
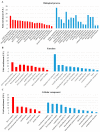
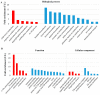
Competitive and naturally occurring yeast killer phenotype is governed by coinfection with dsRNA viruses. Long-term relationship between the host cell and viruses appear to be beneficial and co-adaptive; however, the impact of viral dsRNA on the host gene expression has barely been investigated. Here, we determined the transcriptomic profiles of the host Saccharomyces cerevisiae upon the loss of the M-2 dsRNA alone and the M-2 along with the L-A-lus dsRNAs. We provide a comprehensive study based on the high-throughput RNA-Seq data, Gene Ontology and the analysis of the interaction networks. We identified 486 genes differentially expressed after curing yeast cells of the M-2 dsRNA and 715 genes affected by the elimination of both M-2 and L-A-lus dsRNAs. We report that most of the transcriptional responses induced by viral dsRNAs are moderate. Differently expressed genes are related to ribosome biogenesis, mitochondrial functions, stress response, biosynthesis of lipids and amino acids. Our study also provided insight into the virus-host and virus-virus interplays.
Keywords: RNA-Seq; Saccharomyces cerevisiae; dsRNA viruses; host gene expression.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

