Genetic Breast Cancer Susceptibility Variants and Prognosis in the Prospectively Randomized SUCCESS A Study
- PMID: 28757652
- PMCID: PMC5514410
- DOI: 10.1055/s-0042-113189
Genetic Breast Cancer Susceptibility Variants and Prognosis in the Prospectively Randomized SUCCESS A Study
Abstract
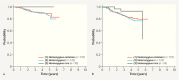
Large-scale genotyping studies have identified over 70 single nucleotide polymorphisms (SNPs) associated with breast cancer (BC) risk. However, knowledge regarding genetic risk factors associated with the prognosis is limited. The aim of this study was therefore to investigate the prognostic effect of nine known breast cancer risk SNPs. BC patients (n = 1687) randomly sampled in an adjuvant, randomized phase III trial (SUCCESS A study) were genotyped for nine BC risk SNPs: rs17468277 (CASP8) , rs2981582 (FGFR2) , rs13281615(8q24), rs3817198 (LSP1) , rs889312 (MAP3K1) , rs3803662 (TOX3) , rs13387042(2q35), rs4973768 (SLC4A7) , rs6504950 (COX11) . Cox proportional hazards models were used to test the SNPs' association with overall survival (OS) and progression-free survival (PFS). Additional analyses were carried out for molecular subgroups. rs3817198 in LSP1 (lymphocyte-specific protein 1) was the only SNP that significantly influenced OS (p = 0.01) and PFS (p < 0.01) in the likelihood ratio test comparing the genetic survival model with the clinical survival model. In the molecular subgroups, triple-negative patients with two minor alleles in rs3817198 had a much better prognosis relative to OS (adjusted HR 0.03; 95% CI 0.002 - 0.279) and PFS (HR 0.09; 95% CI 0.02 - 0.36) than patients with the common alleles. The same effect on PFS was shown for patients with luminal A tumors (HR 0.19; 95% CI 0.05 - 0.84), whereas patients with luminal B tumors had a poorer PFS with two minor alleles (HR 2.13; 95% CI 1.02 - 4.40). The variant in rs3817198 has a prognostic effect particularly in the subgroup of patients with triple-negative BC, suggesting a possible link with immunomodulation and BC.
In verschiedenen großangelegten Studien, in denen umfangreiche Genotypisierungsstudien vorgenommen wurden, wurden bislang über 70 Einzelnukleotid-Polymorphismen (SNPs) identifiziert, die mit einem erhöhten Brustkrebsrisiko einhergehen. Aber das Wissen über die mit der Prognose assoziierten Risikofaktoren wächst weniger schnell. Ziel dieser Studie war es daher, die Auswirkungen von 9 bekannten Brustkrebsrisiko-SNPs auf die Prognose zu untersuchen. In einer adjuvanten randomisierten Phase-III-Studie (SUCCESS A-Studie) wurden Brustkrebspatientinnen (n = 1687) einer Genotypisierung unterzogen. Die Patientinnen waren zuvor nach dem Zufallsprinzip ausgewählt worden. Bei der Genotypisierung standen 9 Brustkrebsrisiko-SNPs im Mittelpunkt: rs17468277 (CASP8) , rs2981582 (FGFR2) , rs13281615(8q24), rs3817198 (LSP1) , rs889312 (MAP3K1) , rs3803662 (TOX3) , rs13387042(2q35), rs4973768 (SLC4A7) , rs6504950 (COX11) . Zur Überprüfung des Zusammenhangs zwischen SNP und Gesamtüberleben (OS) bzw. progressionsfreiem Überleben (PFS) wurde eine Cox-Regressionsanalyse durchgeführt. Molekulare Untergruppen wurden einer weiteren Analyse unterzogen. rs3817198 in LSP1 (Lymphozyten-spezifisches Protein 1) war der einzige SNP, für den im Likelihood-Ratio-Test, der das genetische Überlebensmodell mit dem klinischen Überlebensmodell verglich, eine signifikante Auswirkung auf das Gesamtüberleben (p = 0,01) und das progressionsfreie Überleben (p < 0,01) festgestellt wurde. In den molekularen Untergruppen hatten triple-negative Patientinnen mit 2 seltenen Allelen in rs3817198 eine viel bessere Prognose im Hinblick auf ihr Gesamtüberleben (adjustierte HR 0,03; 95%-KI 0,002 – 0,279) und ihr progressionsfreies Überleben (HR 0,09; 95%-KI 0,02 – 0,36), verglichen mit Patientinnen, welche die häufig vorkommenden Allele aufwiesen. Dieselbe Auswirkung auf das progressionsfreie Überleben fand sich auch bei Patientinnen mit Brustkrebs vom Luminal-A-Typ (HR 0,19; 95%-KI 0,05 – 0,84), während Patientinnen mit Brustkrebs vom Typ Luminal-B und 2 seltenen Allelen ein geringeres progressionsfreies Überleben aufwiesen (HR 2,13; 95%-KI 1,02 – 4,40). Die Variante in rs3817198 hatte besonders auf die Untergruppe von Patientinnen mit triple-negativem Brustkrebs eine prognostische Auswirkung, was auf eine mögliche Assoziation zwischen Immunmodulation und Brustkrebs hindeutet.
Keywords: LSP1; SNP; TNBC; breast cancer; prognosis.
Conflict of interest statement
Figures
Similar articles
-
The role of genetic breast cancer susceptibility variants as prognostic factors.Hum Mol Genet. 2012 Sep 1;21(17):3926-39. doi: 10.1093/hmg/dds159. Epub 2012 Apr 24. Hum Mol Genet. 2012. PMID: 22532573 Free PMC article.
-
Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium.J Natl Cancer Inst. 2011 Aug 17;103(16):1252-63. doi: 10.1093/jnci/djr265. Epub 2011 Jul 26. J Natl Cancer Inst. 2011. PMID: 21791674 Free PMC article.
-
Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study.Breast Cancer Res. 2010;12(6):R110. doi: 10.1186/bcr2797. Epub 2010 Dec 31. Breast Cancer Res. 2010. PMID: 21194473 Free PMC article.
-
Low-penetrance susceptibility variants and postmenopausal oestrogen receptor positive breast cancer.J Genet. 2020;99:15. J Genet. 2020. PMID: 32366738
-
Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies.Breast Cancer Res Treat. 2011 Jun;127(2):309-24. doi: 10.1007/s10549-011-1459-5. Epub 2011 Mar 29. Breast Cancer Res Treat. 2011. PMID: 21445572 Review.
Cited by
-
Update Breast Cancer 2024 Part 2 - Patients with Early Stage Breast Cancer.Geburtshilfe Frauenheilkd. 2025 May 15;85(5):493-506. doi: 10.1055/a-2533-2783. eCollection 2025 May. Geburtshilfe Frauenheilkd. 2025. PMID: 40386499 Free PMC article.
-
Mutations in BRCA1/2 and Other Panel Genes in Patients With Metastatic Breast Cancer -Association With Patient and Disease Characteristics and Effect on Prognosis.J Clin Oncol. 2021 May 20;39(15):1619-1630. doi: 10.1200/JCO.20.01200. Epub 2021 Mar 29. J Clin Oncol. 2021. PMID: 33780288 Free PMC article.
-
Cost effectiveness of bilateral risk-reducing mastectomy and salpingo-oophorectomy.Eur J Med Res. 2019 Sep 14;24(1):32. doi: 10.1186/s40001-019-0391-8. Eur J Med Res. 2019. PMID: 31521205 Free PMC article.
-
Update Breast Cancer 2018 (Part 1) - Primary Breast Cancer and Biomarkers.Geburtshilfe Frauenheilkd. 2018 Mar;78(3):237-245. doi: 10.1055/s-0044-101613. Epub 2018 Mar 21. Geburtshilfe Frauenheilkd. 2018. PMID: 29576629 Free PMC article.
-
Association of the Estrogen Receptor 1 Polymorphisms rs2046210 and rs9383590 with the Risk, Age at Onset and Prognosis of Breast Cancer.Cells. 2023 Feb 4;12(4):515. doi: 10.3390/cells12040515. Cells. 2023. PMID: 36831182 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous