An iridium(iii)-based irreversible protein-protein interaction inhibitor of BRD4 as a potent anticancer agent
- PMID: 28757943
- PMCID: PMC5510529
- DOI: 10.1039/c5sc02321a
An iridium(iii)-based irreversible protein-protein interaction inhibitor of BRD4 as a potent anticancer agent
Abstract
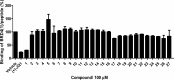
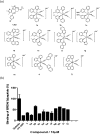
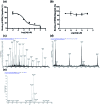
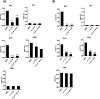
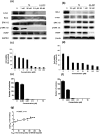
Bromodomain-containing protein 4 (BRD4) has recently emerged as an attractive epigenetic target for anticancer therapy. In this study, an iridium(iii) complex is reported as the first metal-based, irreversible inhibitor of BRD4. Complex 1a is able to antagonize the BRD4-acetylated histone protein-protein interaction (PPI) in vitro, and to bind BRD4 and down-regulate c-myc oncogenic expression in cellulo. Chromatin immunoprecipitation (ChIP) analysis revealed that 1a could modulate the interaction between BRD4 and chromatin in melanoma cells, particular at the MYC promoter. Finally, the complex showed potent activity against melanoma xenografts in an in vivo mouse model. To our knowledge, this is the first report of a Group 9 metal complex inhibiting the PPI of a member of the bromodomain and extraterminal domain (BET) family. We envision that complex 1a may serve as a useful scaffold for the development of more potent epigenetic agents against cancers such as melanoma.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

