Constructing real-time, wash-free, and reiterative sensors for cell surface proteins using binding-induced dynamic DNA assembly
- PMID: 28757954
- PMCID: PMC5514693
- DOI: 10.1039/c5sc01870f
Constructing real-time, wash-free, and reiterative sensors for cell surface proteins using binding-induced dynamic DNA assembly
Abstract
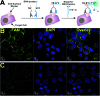
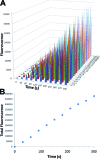
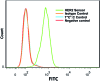
Cell surface proteins are an important class of biomarkers for fundamental biological research and for disease diagnostics and treatment. In this communication, we report a universal strategy to construct sensors that can achieve rapid imaging of cell surface proteins without any separation by using binding-induced dynamic DNA assembly. As a proof-of-principle, we developed a real-time and wash-free sensor for an important breast cancer biomarker, human epidermal growth factor receptor-2 (HER2). We then demonstrated that this sensor could be used for imaging and sensing HER2 on both fixed and live breast cancer cells. Additionally, we have also incorporated toehold-mediated DNA strand displacement reactions into the HER2 sensor, which allows for reiterating (switching on/off) fluorescence signals for HER2 from breast cancer cells in real-time.
Figures


Similar articles
-
Regulation of DNA Strand Displacement Using an Allosteric DNA Toehold.J Am Chem Soc. 2016 Oct 26;138(42):14076-14082. doi: 10.1021/jacs.6b08794. Epub 2016 Oct 13. J Am Chem Soc. 2016. PMID: 27704809
-
Toehold enabling stem-loop inspired hemiduplex probe with enhanced sensitivity and sequence-specific detection of tumor DNA in serum.Biosens Bioelectron. 2016 Aug 15;82:32-9. doi: 10.1016/j.bios.2016.03.054. Epub 2016 Mar 23. Biosens Bioelectron. 2016. PMID: 27040528
-
Amplified fluorescence imaging of HER2 dimerization on cancer cells by using a co-localization triggered DNA nanoassembly.Mikrochim Acta. 2019 Jun 13;186(7):439. doi: 10.1007/s00604-019-3549-8. Mikrochim Acta. 2019. PMID: 31197538
-
Can we define breast cancer HER2 status by liquid biopsy?Int Rev Cell Mol Biol. 2023;381:23-56. doi: 10.1016/bs.ircmb.2023.07.003. Epub 2023 Sep 4. Int Rev Cell Mol Biol. 2023. PMID: 37739483 Review.
-
Principles and Applications of Nucleic Acid Strand Displacement Reactions.Chem Rev. 2019 May 22;119(10):6326-6369. doi: 10.1021/acs.chemrev.8b00580. Epub 2019 Feb 4. Chem Rev. 2019. PMID: 30714375 Review.
Cited by
-
Dynamically elongated associative toehold for tuning DNA circuit kinetics and thermodynamics.Nucleic Acids Res. 2021 May 7;49(8):4258-4265. doi: 10.1093/nar/gkab212. Nucleic Acids Res. 2021. PMID: 33849054 Free PMC article.
-
A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers.Chem Sci. 2020 Oct 12;11(44):12157-12164. doi: 10.1039/d0sc03920a. Chem Sci. 2020. PMID: 34123223 Free PMC article.
-
A photocleavable peptide-tagged mass probe for chemical mapping of epidermal growth factor receptor 2 (HER2) in human cancer cells.Chem Sci. 2020 Sep 30;11(41):11298-11306. doi: 10.1039/d0sc04481d. Chem Sci. 2020. PMID: 34094372 Free PMC article.
-
Cofactor-assisted three-way DNA junction-driven strand displacement.RSC Adv. 2021 Sep 10;11(48):30377-30382. doi: 10.1039/d1ra05242j. eCollection 2021 Sep 6. RSC Adv. 2021. PMID: 35480263 Free PMC article.
-
A novel dual-release scaffold for fluorescent labels improves cyclic immunofluorescence.RSC Chem Biol. 2024 May 28;5(7):684-690. doi: 10.1039/d4cb00007b. eCollection 2024 Jul 3. RSC Chem Biol. 2024. PMID: 38966675 Free PMC article.
References
-
- Zhang H., Li F., Dever B., Li X. F., le X. C., Zhang D. Y., Seelig G., Jung C., Ellington A. D. Chem. Rev. Nat. Chem. Acc. Chem. Res. 2013;2011;2014;113347:2812–2841. 103–113, 1825–1835. - PubMed
-
- Dirks R. M., Pierce N. A., Choi H. M., Beck V. A., Pierce N. A., Wu Z., Liu G. Q., Yang X. L., Jiang J. H. Proc. Natl. Acad. Sci. U. S. A. ACS Nano. J. Am. Chem. Soc. 2004;2014;2015;1018137:15274–15278. 4284–4294, 6829–6836.
-
- Yin P., Choi H. M. T., Calvert C. R., Pierce N. A., Li B., Ellington A. D., Chen X., Jiang Y. S., Li B., Milligan J. N., Bhadra S., Ellington A. D., Wu C., Cansiz S., Zhang L. Nature. Nucleic Acids Res. J. Am. Chem. Soc. J. Am. Chem. Soc. 2008;2011;2013;2015;45139135137:318–322. e110, 7430–7433, 4900–4903. - PubMed
-
- Li F., Zhang H., Wang Z., Li X., Li X. F., le X. C., Li F., Zhang H., Lai C., Li X. F., le X. C., Zhang H., Li F., Dever B., Wang C., Li X. F., le X. C., Li F., Lin Y., le X. C. J. Am. Chem. Soc. Angew. Chem., Int. Ed. Angew. Chem., Int. Ed. Anal. Chem. 2013;2012;2013;2013;135515285:2443–2446. 9317–9320, 10698–106705, 10835–10841. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

