A Different Microbiome Gene Repertoire in the Airways of Cystic Fibrosis Patients with Severe Lung Disease
- PMID: 28758937
- PMCID: PMC5578044
- DOI: 10.3390/ijms18081654
A Different Microbiome Gene Repertoire in the Airways of Cystic Fibrosis Patients with Severe Lung Disease
Abstract
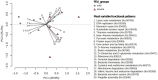
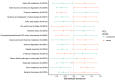
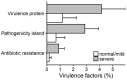
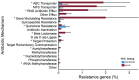
In recent years, next-generation sequencing (NGS) was employed to decipher the structure and composition of the microbiota of the airways in cystic fibrosis (CF) patients. However, little is still known about the overall gene functions harbored by the resident microbial populations and which specific genes are associated with various stages of CF lung disease. In the present study, we aimed to identify the microbial gene repertoire of CF microbiota in twelve patients with severe and normal/mild lung disease by performing sputum shotgun metagenome sequencing. The abundance of metabolic pathways encoded by microbes inhabiting CF airways was reconstructed from the metagenome. We identified a set of metabolic pathways differently distributed in patients with different pulmonary function; namely, pathways related to bacterial chemotaxis and flagellar assembly, as well as genes encoding efflux-mediated antibiotic resistance mechanisms and virulence-related genes. The results indicated that the microbiome of CF patients with low pulmonary function is enriched in virulence-related genes and in genes encoding efflux-mediated antibiotic resistance mechanisms. Overall, the microbiome of severely affected adults with CF seems to encode different mechanisms for the facilitation of microbial colonization and persistence in the lung, consistent with the characteristics of multidrug-resistant microbial communities that are commonly observed in patients with severe lung disease.
Keywords: bioinformatics; cystic fibrosis; lung disease; lung microbiome; metabolic pathways; shotgun metagenomics; virulence genes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures


References
-
- Cystic Fibrosis Foundation . Patient Registry Annual Data Report 2015. Cystic Fibrosis Foundation; Bethesda, MD, USA: 2016.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

