Non-Targeted Metabolomics Analysis of Golden Retriever Muscular Dystrophy-Affected Muscles Reveals Alterations in Arginine and Proline Metabolism, and Elevations in Glutamic and Oleic Acid In Vivo
- PMID: 28758940
- PMCID: PMC5618323
- DOI: 10.3390/metabo7030038
Non-Targeted Metabolomics Analysis of Golden Retriever Muscular Dystrophy-Affected Muscles Reveals Alterations in Arginine and Proline Metabolism, and Elevations in Glutamic and Oleic Acid In Vivo
Abstract
Background: Like Duchenne muscular dystrophy (DMD), the Golden Retriever Muscular Dystrophy (GRMD) dog model of DMD is characterized by muscle necrosis, progressive paralysis, and pseudohypertrophy in specific skeletal muscles. This severe GRMD phenotype includes moderate atrophy of the biceps femoris (BF) as compared to unaffected normal dogs, while the long digital extensor (LDE), which functions to flex the tibiotarsal joint and serves as a digital extensor, undergoes the most pronounced atrophy. A recent microarray analysis of GRMD identified alterations in genes associated with lipid metabolism and energy production.
Methods: We, therefore, undertook a non-targeted metabolomics analysis of the milder/earlier stage disease GRMD BF muscle versus the more severe/chronic LDE using GC-MS to identify underlying metabolic defects specific for affected GRMD skeletal muscle.
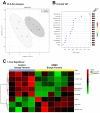
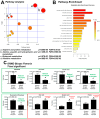
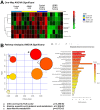
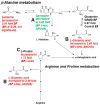
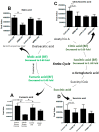
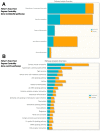
Results: Untargeted metabolomics analysis of moderately-affected GRMD muscle (BF) identified eight significantly altered metabolites, including significantly decreased stearamide (0.23-fold of controls, p = 2.89 × 10-3), carnosine (0.40-fold of controls, p = 1.88 × 10-2), fumaric acid (0.40-fold of controls, p = 7.40 × 10-4), lactamide (0.33-fold of controls, p = 4.84 × 10-2), myoinositol-2-phosphate (0.45-fold of controls, p = 3.66 × 10-2), and significantly increased oleic acid (1.77-fold of controls, p = 9.27 × 10-2), glutamic acid (2.48-fold of controls, p = 2.63 × 10-2), and proline (1.73-fold of controls, p = 3.01 × 10-2). Pathway enrichment analysis identified significant enrichment for arginine/proline metabolism (p = 5.88 × 10-4, FDR 4.7 × 10-2), where alterations in L-glutamic acid, proline, and carnosine were found. Additionally, multiple Krebs cycle intermediates were significantly decreased (e.g., malic acid, fumaric acid, citric/isocitric acid, and succinic acid), suggesting that altered energy metabolism may be underlying the observed GRMD BF muscle dysfunction. In contrast, two pathways, inosine-5'-monophosphate (VIP Score 3.91) and 3-phosphoglyceric acid (VIP Score 3.08) mainly contributed to the LDE signature, with two metabolites (phosphoglyceric acid and inosine-5'-monophosphate) being significantly decreased. When the BF and LDE were compared, the most significant metabolite was phosphoric acid, which was significantly less in the GRMD BF compared to control and GRMD LDE groups.
Conclusions: The identification of elevated BF oleic acid (a long-chain fatty acid) is consistent with recent microarray studies identifying altered lipid metabolism genes, while alterations in arginine and proline metabolism are consistent with recent studies identifying elevated L-arginine in DMD patient sera as a biomarker of disease. Together, these studies demonstrate muscle-specific alterations in GRMD-affected muscle, which illustrate previously unidentified metabolic changes.
Keywords: Duchenne muscular dystrophy; golden retriever muscular dystrophy; metabolism; non-targeted metabolomics; skeletal muscle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

