SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling
- PMID: 28759002
- PMCID: PMC5712491
- DOI: 10.1038/ni.3809
SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling
Abstract
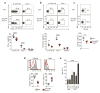
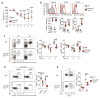
Among the features that distinguish type 1 innate lymphoid cells (ILC1s) from natural killer (NK) cells is a gene signature indicative of 'imprinting' by cytokines of the TGF-β family. We studied mice in which ILC1s and NK cells lacked SMAD4, a signal transducer that facilitates the canonical signaling pathway common to all cytokines of the TGF-β family. While SMAD4 deficiency did not affect ILC1 differentiation, NK cells unexpectedly acquired an ILC1-like gene signature and were unable to control tumor metastasis or viral infection. Mechanistically, SMAD4 restrained non-canonical TGF-β signaling mediated by the cytokine receptor TGFβR1 in NK cells. NK cells from a SMAD4-deficient person affected by polyposis were also hyper-responsive to TGF-β. These results identify SMAD4 as a previously unknown regulator that restricts non-canonical TGF-β signaling in NK cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



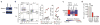
References
-
- Klose CSN, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

