Implications of human genetic variation in CRISPR-based therapeutic genome editing
- PMID: 28759051
- PMCID: PMC5749234
- DOI: 10.1038/nm.4377
Implications of human genetic variation in CRISPR-based therapeutic genome editing
Abstract
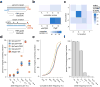
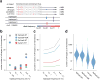
CRISPR-Cas genome-editing methods hold immense potential as therapeutic tools to fix disease-causing mutations at the level of DNA. In contrast to typical drug development strategies aimed at targets that are highly conserved among individual patients, treatment at the genomic level must contend with substantial inter-individual natural genetic variation. Here we analyze the recently released ExAC and 1000 Genomes data sets to determine how human genetic variation impacts target choice for Cas endonucleases in the context of therapeutic genome editing. We find that this genetic variation confounds the target sites of certain Cas endonucleases more than others, and we provide a compendium of guide RNAs predicted to have high efficacy in diverse patient populations. For further analysis, we focus on 12 therapeutically relevant genes and consider how genetic variation affects off-target candidates for these loci. Our analysis suggests that, in large populations of individuals, most candidate off-target sites will be rare, underscoring the need for prescreening of patients through whole-genome sequencing to ensure safety. This information can be integrated with empirical methods for guide RNA selection into a framework for designing CRISPR-based therapeutics that maximizes efficacy and safety across patient populations.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

