Transcriptome analysis of the tea oil camellia (Camellia oleifera) reveals candidate drought stress genes
- PMID: 28759610
- PMCID: PMC5536306
- DOI: 10.1371/journal.pone.0181835
Transcriptome analysis of the tea oil camellia (Camellia oleifera) reveals candidate drought stress genes
Abstract
Background: The tea-oil camellia (Camellia oleifera) is the most important oil plant in southern China, and has a strong resistance to drought and barren soil. Understanding the molecular mechanisms of drought tolerance would greatly promote its cultivation and molecular breeding.
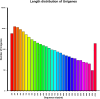
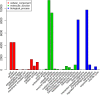
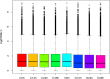
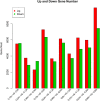
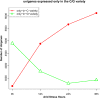
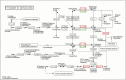
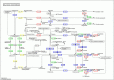
Results: In total, we obtained 76,585 unigenes with an average length of 810 bp and an N50 of 1,092 bp. We mapped all the unigenes to the NCBI 'nr' (non-redundant), SwissProt, KEGG, and clusters of orthologous groups (COG) databases, where 52,531 (68.6%) unigenes were functionally annotated. According to the annotation, 46,171 (60.8%) unigenes belong to 338 KEGG pathways. We identified a series of unigenes that are related to the synthesis and regulation of abscisic acid (ABA), the activity of protective enzymes, vitamin B6 metabolism, the metabolism of osmolytes, and pathways related to the biosynthesis of secondary metabolites. After exposed to drought for 12 hours, the number of differentially-expressed genes (DEGs) between treated plants and control plants increased in the G4 cultivar, while there was no significant increase in the drought-tolerant C3 cultivar. DEGs associated with drought stress responsive pathways were identified by KEGG pathway enrichment analysis. Moreover, we found 789 DEGs related to transcription factors. Finally, according to the results of qRT-PCR, the expression levels of the 20 unigenes tested were consistent with the results of next-generation sequencing.
Conclusions: In the present study, we identified a large set of cDNA unigenes from C. oleifera annotated using public databases. Further studies of DEGs involved in metabolic pathways related to drought stress and transcription will facilitate the discovery of novel genes involved in resistance to drought stress in this commercially important plant.
Conflict of interest statement
Figures






References
-
- Ma JL, Ye H, Rui YK, Chen GC, Zhang NY. Fatty acid composition of Camellia oleifera oil. J Verbrauch Lebensm. 2011; 6(1): 9–12.
-
- Xia EH, Jiang JJ, Huang H, Zhang LP, Zhang HB, Gao LZ. Transcriptome analysis of the oil-rich tea plant, Camellia oleifera, reveals candidate genes related to lipid metabolism. PLoS One. 2014; 9(8): e104150 doi: 10.1371/journal.pone.0104150 - DOI - PMC - PubMed
-
- Lee CP, Yen GC. Antioxidant activity and bioactive compounds of tea seed (Camellia oleifera Abel.) oil. J Agric Food Chem. 2006; 54(3): 779–784. doi: 10.1021/jf052325a - DOI - PubMed
-
- Chen YF, Yang CH, Chang MS, Ciou YP, Huang YC. Foam properties and detergent abilities of the saponins from Camellia oleifera. Int J Mol Sci. 2010; 11(11): 4417–4425. doi: 10.3390/ijms11114417 - DOI - PMC - PubMed
-
- Zhang XF, Yang SL, Han YY, Zhao L, Lu GL, Xia T, et al. Qualitative and quantitative analysis of triterpene saponins from tea seed pomace (Camellia oleifera Abel.) and their activities against bacteria and fungi. Molecules. 2014; 19(6): 7568–7580. doi: 10.3390/molecules19067568 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

