Rare HIV-1 transmitted/founder lineages identified by deep viral sequencing contribute to rapid shifts in dominant quasispecies during acute and early infection
- PMID: 28759651
- PMCID: PMC5552316
- DOI: 10.1371/journal.ppat.1006510
Rare HIV-1 transmitted/founder lineages identified by deep viral sequencing contribute to rapid shifts in dominant quasispecies during acute and early infection
Erratum in
-
Correction: Rare HIV-1 transmitted/founder lineages identified by deep viral sequencing contribute to rapid shifts in dominant quasispecies during acute and early infection.PLoS Pathog. 2017 Sep 14;13(9):e1006620. doi: 10.1371/journal.ppat.1006620. eCollection 2017 Sep. PLoS Pathog. 2017. PMID: 28910384 Free PMC article.
Abstract
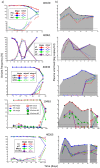
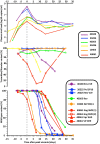
In order to inform the rational design of HIV-1 preventive and cure interventions it is critical to understand the events occurring during acute HIV-1 infection (AHI). Using viral deep sequencing on six participants from the early capture acute infection RV217 cohort, we have studied HIV-1 evolution in plasma collected twice weekly during the first weeks following the advent of viremia. The analysis of infections established by multiple transmitted/founder (T/F) viruses revealed novel viral profiles that included: a) the low-level persistence of minor T/F variants, b) the rapid replacement of the major T/F by a minor T/F, and c) an initial expansion of the minor T/F followed by a quick collapse of the same minor T/F to low frequency. In most participants, cytotoxic T-lymphocyte (CTL) escape was first detected at the end of peak viremia downslope, proceeded at higher rates than previously measured in HIV-1 infection, and usually occurred through the exploration of multiple mutational pathways within an epitope. The rapid emergence of CTL escape variants suggests a strong and early CTL response. Minor T/F viral strains can contribute to rapid and varied profiles of HIV-1 quasispecies evolution during AHI. Overall, our results demonstrate that early, deep, and frequent sampling is needed to investigate viral/host interaction during AHI, which could help identify prerequisites for prevention and cure of HIV-1 infection.
Conflict of interest statement
GHK is currently an employee of GSK Vaccines, Rockville, MD. The authors have declared that no competing interests exist.
Figures



References
-
- (UNAIDS) JUNPoHA. The gap report. Geneva, Switzerland2014.
-
- Palella FJ Jr., Deloria-Knoll M, Chmiel JS, Moorman AC, Wood KC, Greenberg AE, et al. Survival benefit of initiating antiretroviral therapy in HIV-infected persons in different CD4+ cell strata. Ann Intern Med. 2003;138(8):620–6. . - PubMed
-
- Fauci AS, Marston HD. Focusing to achieve a world without AIDS. JAMA. 2015;313(4):357–8. doi: 10.1001/jama.2014.17454 . - DOI - PubMed
-
- Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med. 2012;2(11). doi: 10.1101/cshperspect.a006965 ; PubMed Central PMCID: PMCPMC3543106. - DOI - PMC - PubMed
-
- Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, et al. Prospective Study of Acute HIV-1 Infection in Adults in East Africa and Thailand. N Engl J Med. 2016;374(22):2120–30. doi: 10.1056/NEJMoa1508952 ; PubMed Central PMCID: PMCPMC5111628. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

