Mitotic progression following DNA damage enables pattern recognition within micronuclei
- PMID: 28759889
- PMCID: PMC5857357
- DOI: 10.1038/nature23470
Mitotic progression following DNA damage enables pattern recognition within micronuclei
Abstract
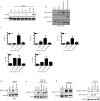
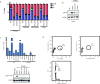
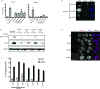
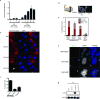
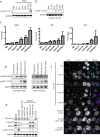
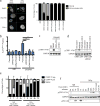
Inflammatory gene expression following genotoxic cancer therapy is well documented, yet the events underlying its induction remain poorly understood. Inflammatory cytokines modify the tumour microenvironment by recruiting immune cells and are critical for both local and systemic (abscopal) tumour responses to radiotherapy. A poorly understood feature of these responses is the delayed onset (days), in contrast to the acute DNA-damage responses that occur in minutes to hours. Such dichotomous kinetics implicate additional rate-limiting steps that are essential for DNA-damage-induced inflammation. Here we show that cell cycle progression through mitosis following double-stranded DNA breaks leads to the formation of micronuclei, which precede activation of inflammatory signalling and are a repository for the pattern-recognition receptor cyclic GMP-AMP synthase (cGAS). Inhibiting progression through mitosis or loss of pattern recognition by stimulator of interferon genes (STING)-cGAS impaired interferon signalling. Moreover, STING loss prevented the regression of abscopal tumours in the context of ionizing radiation and immune checkpoint blockade in vivo. These findings implicate temporal modulation of the cell cycle as an important consideration in the context of therapeutic strategies that combine genotoxic agents with immune checkpoint blockade.
Figures




Comment in
-
cGAS Micro-Manages Genotoxic Stress.Immunity. 2017 Oct 17;47(4):616-617. doi: 10.1016/j.immuni.2017.09.020. Immunity. 2017. PMID: 29045895
References
-
- Cohen Jonathan E, Bernhard EJ, McKenna WG. How does radiation kill cells? Current Opinion in Chemical Biology. 1999;3:77–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

