The three-dimensional genome organization of Drosophila melanogaster through data integration
- PMID: 28760140
- PMCID: PMC5576134
- DOI: 10.1186/s13059-017-1264-5
The three-dimensional genome organization of Drosophila melanogaster through data integration
Abstract
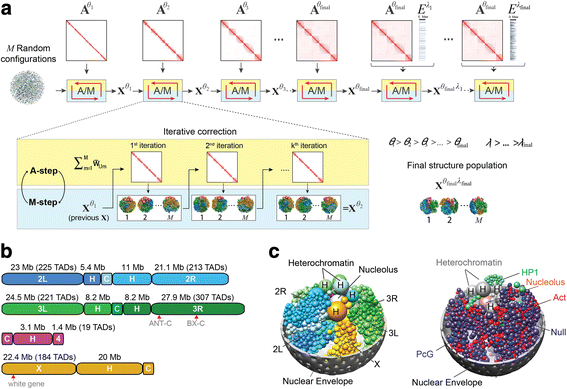
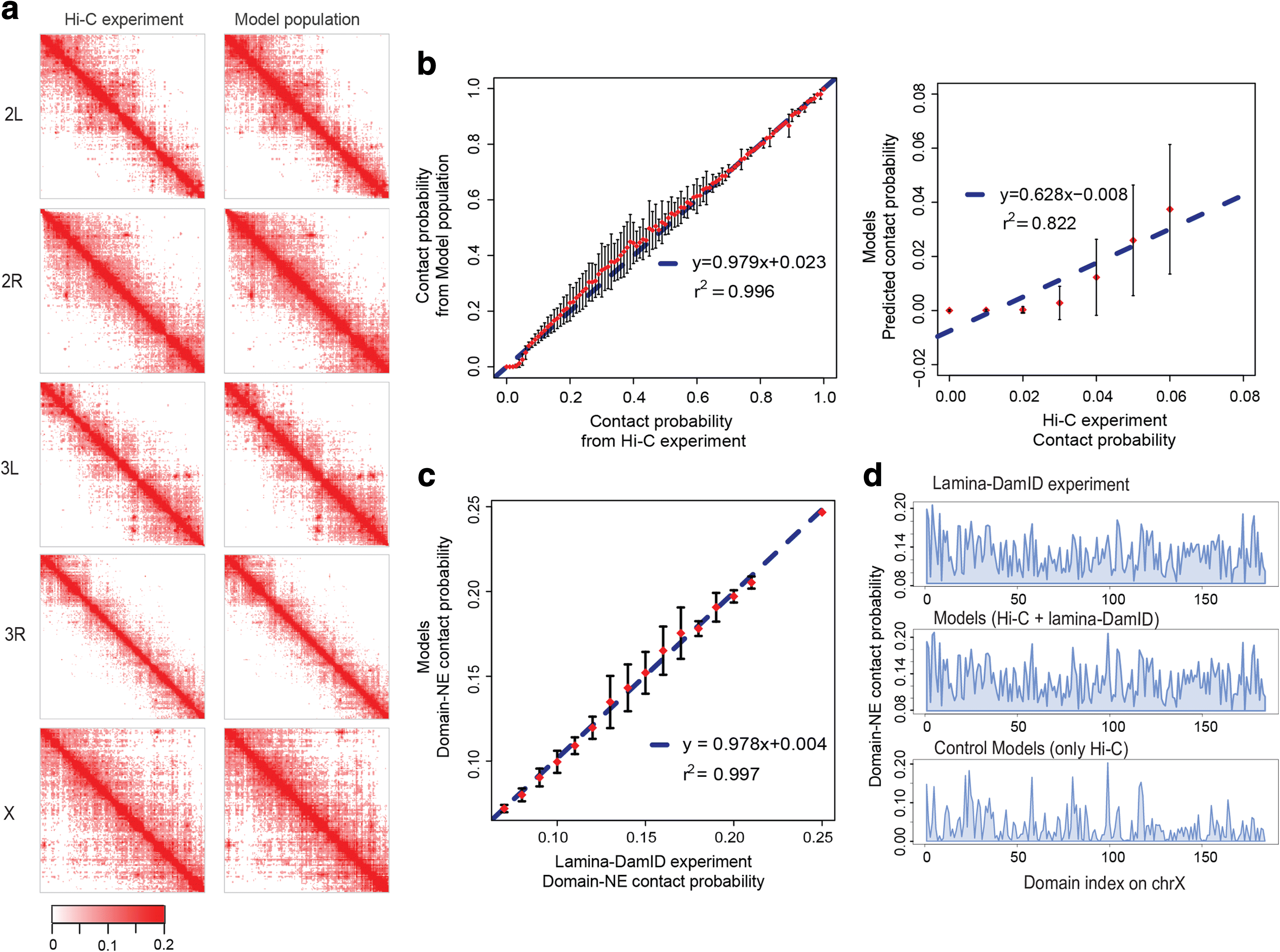
Background: Genome structures are dynamic and non-randomly organized in the nucleus of higher eukaryotes. To maximize the accuracy and coverage of three-dimensional genome structural models, it is important to integrate all available sources of experimental information about a genome's organization. It remains a major challenge to integrate such data from various complementary experimental methods. Here, we present an approach for data integration to determine a population of complete three-dimensional genome structures that are statistically consistent with data from both genome-wide chromosome conformation capture (Hi-C) and lamina-DamID experiments.
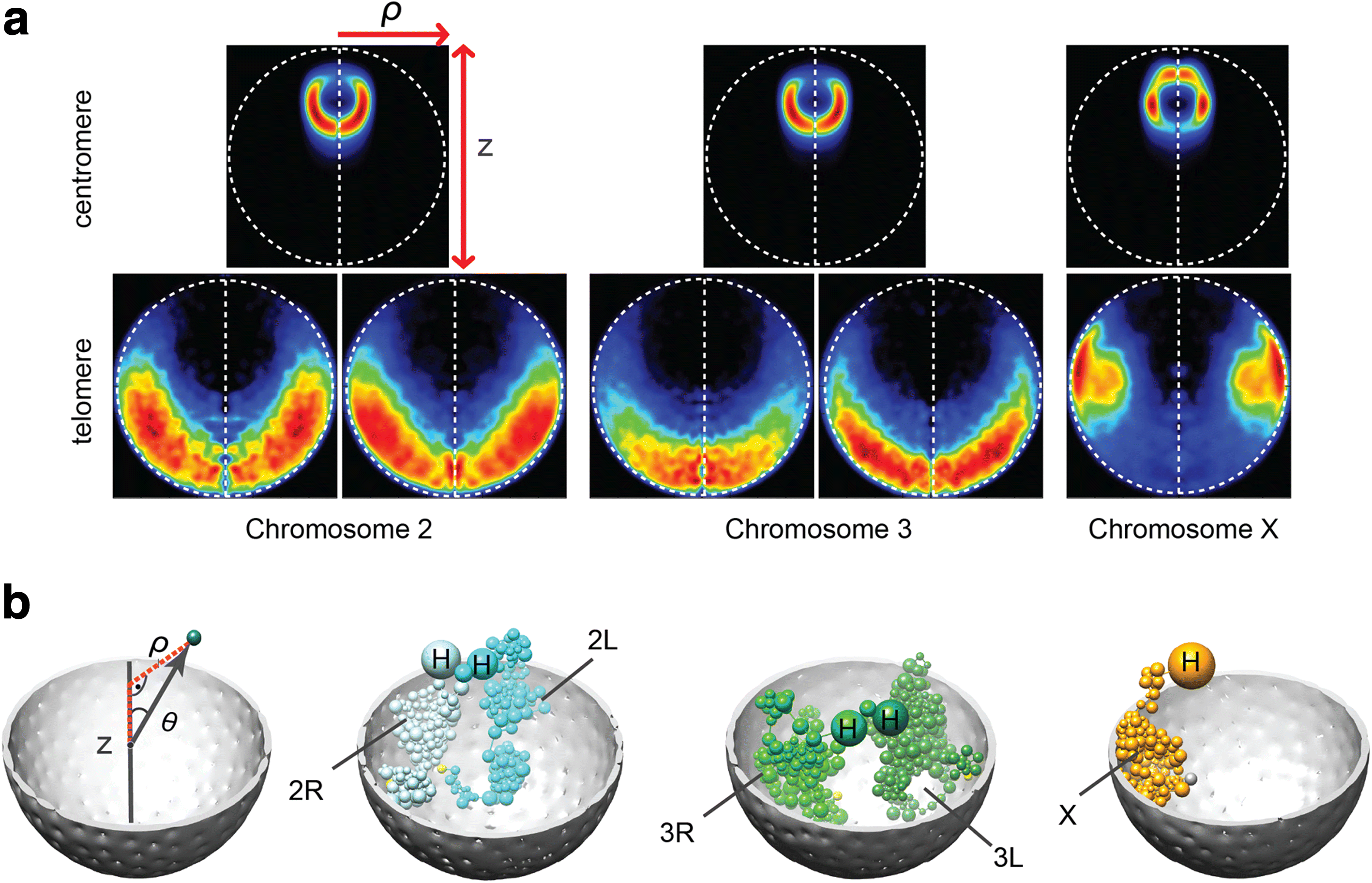
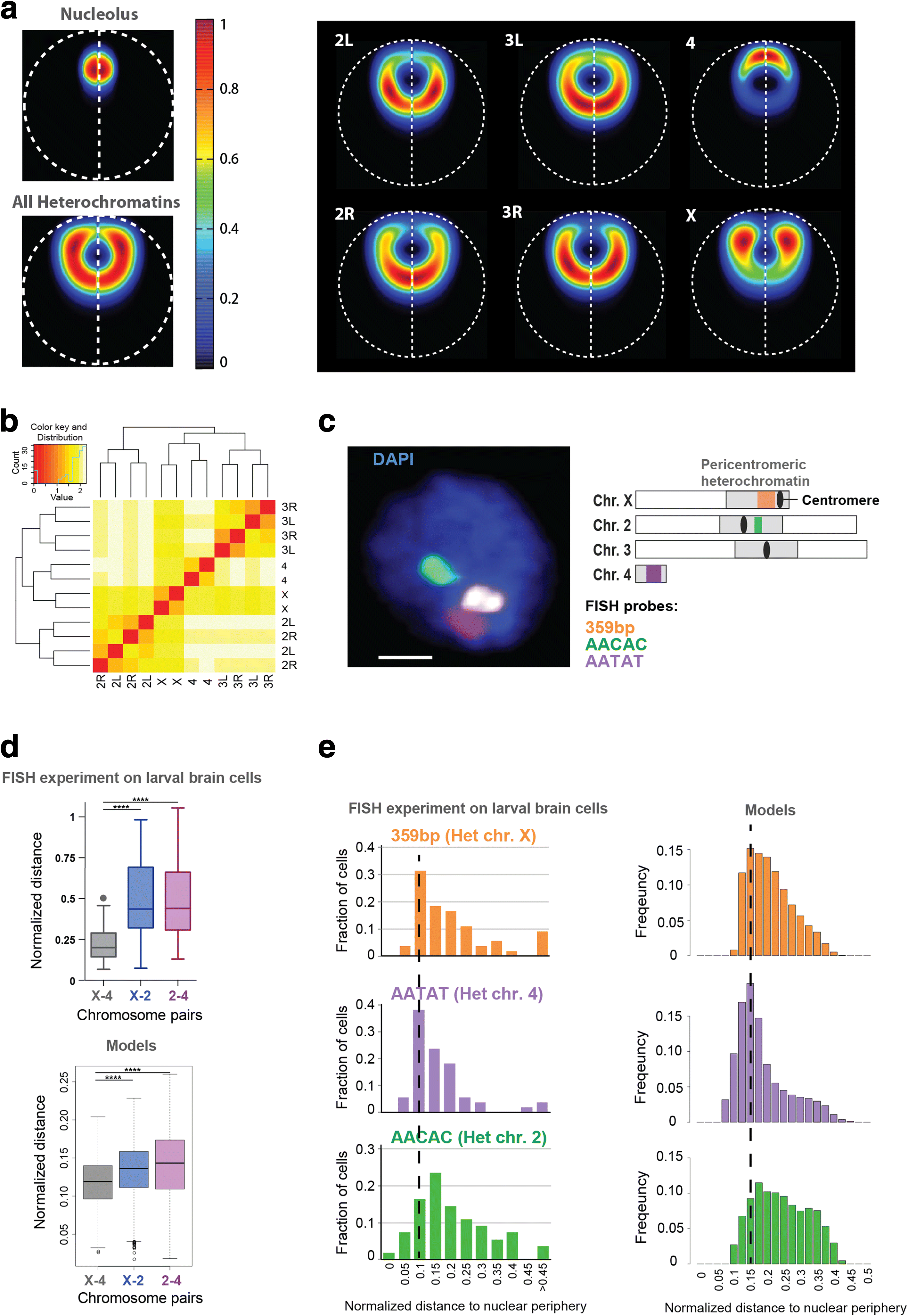
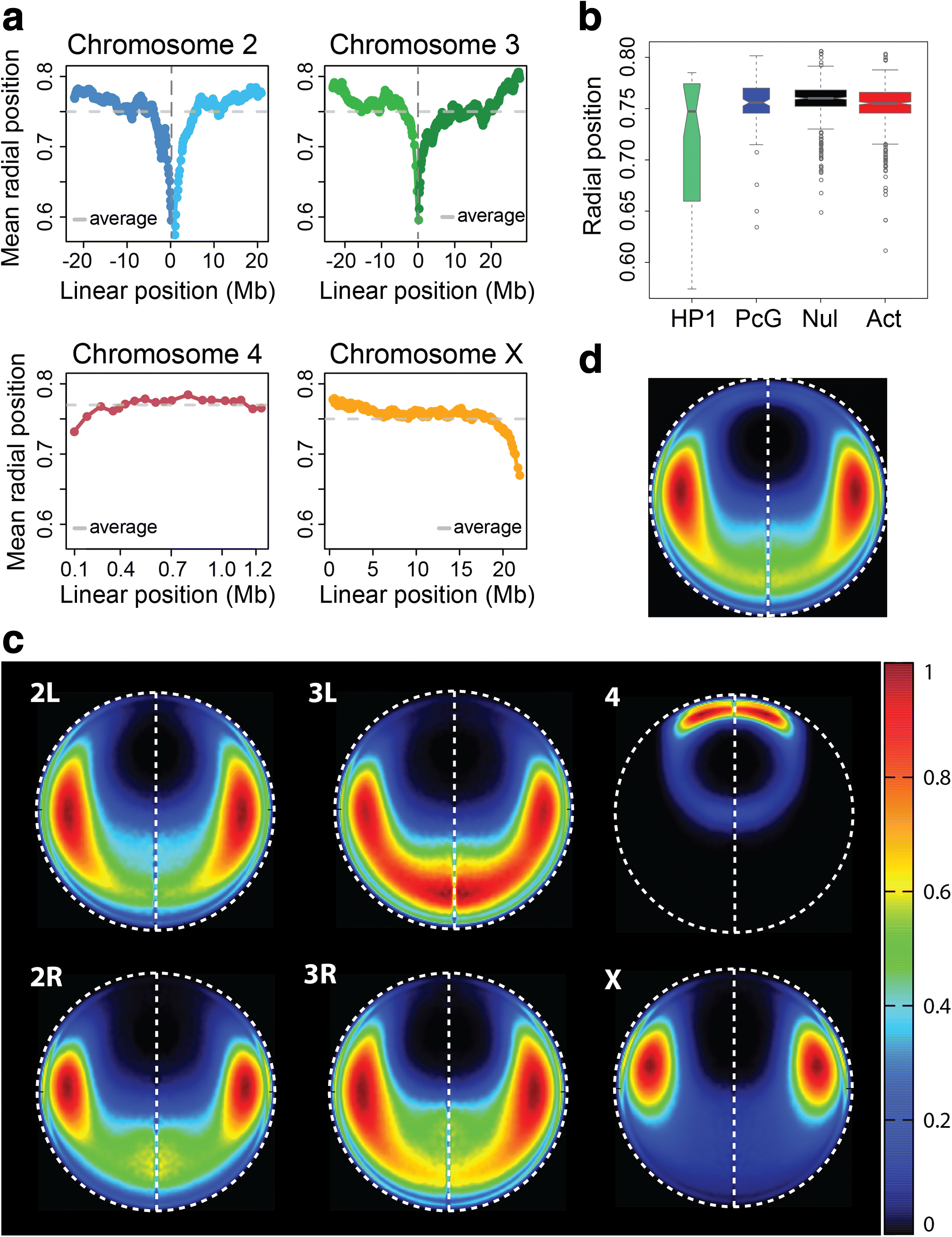
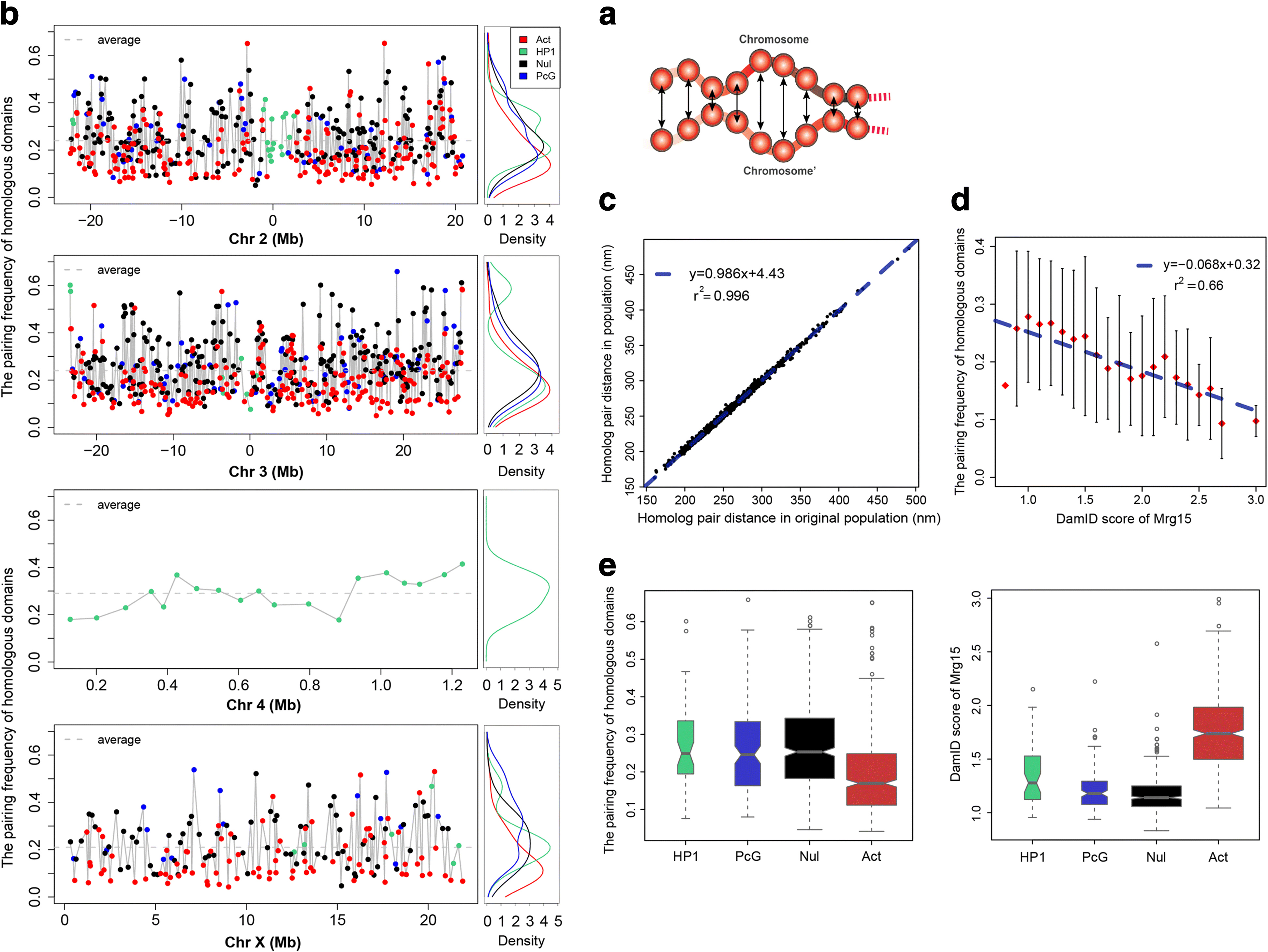
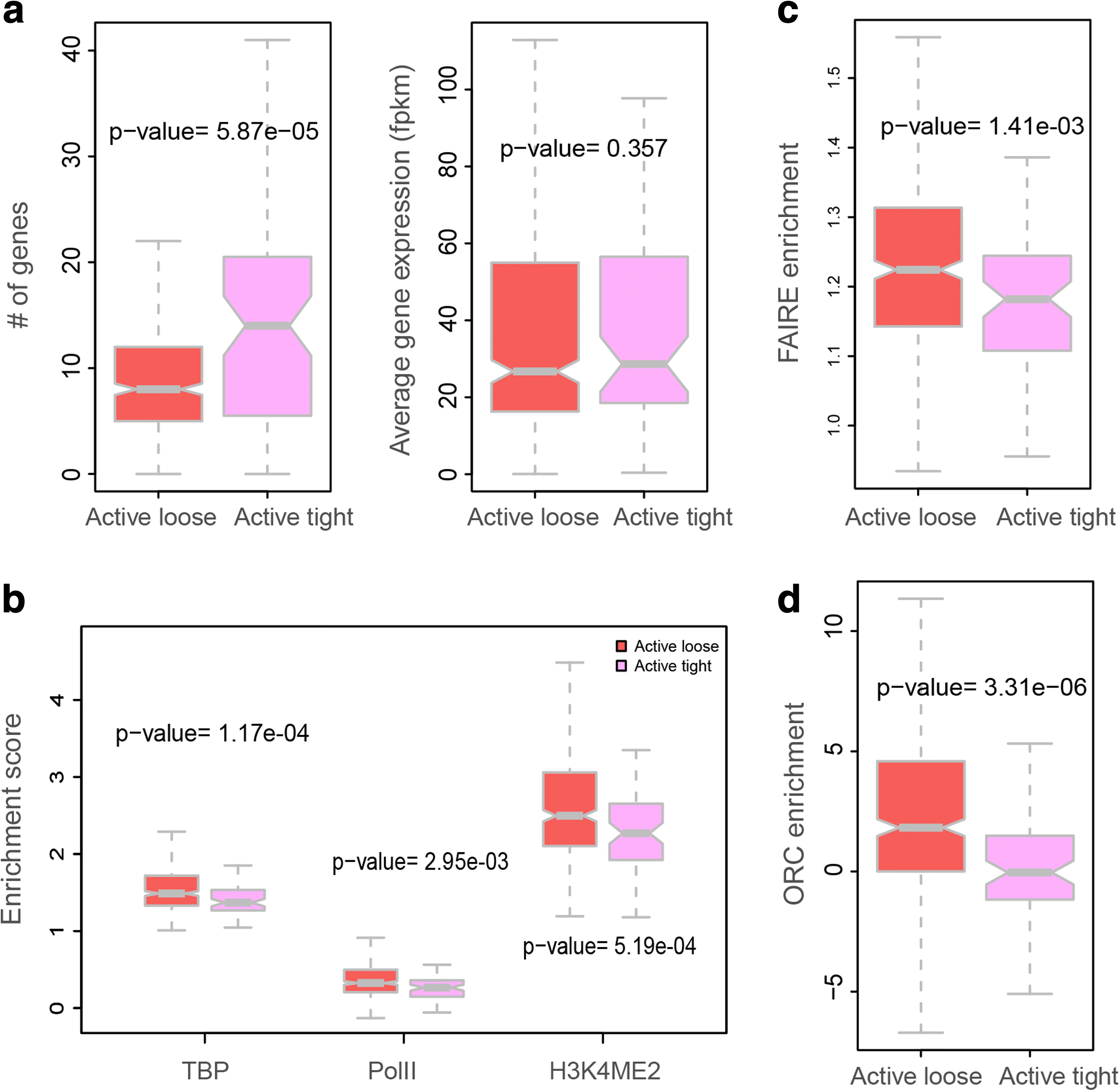
Results: Our structures resolve the genome at the resolution of topological domains, and reproduce simultaneously both sets of experimental data. Importantly, this data deconvolution framework allows for structural heterogeneity between cells, and hence accounts for the expected plasticity of genome structures. As a case study we choose Drosophila melanogaster embryonic cells, for which both data types are available. Our three-dimensional genome structures have strong predictive power for structural features not directly visible in the initial data sets, and reproduce experimental hallmarks of the D. melanogaster genome organization from independent and our own imaging experiments. Also they reveal a number of new insights about genome organization and its functional relevance, including the preferred locations of heterochromatic satellites of different chromosomes, and observations about homologous pairing that cannot be directly observed in the original Hi-C or lamina-DamID data.
Conclusions: Our approach allows systematic integration of Hi-C and lamina-DamID data for complete three-dimensional genome structure calculation, while also explicitly considering genome structural variability.
Keywords: 3D genome structure; Data integration; Drosophila melanogaster; Heterochromatin; Hi-C; Higher order genome organization; Homologous pairing; Lamina-DamID; Population-based modeling.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. - DOI - PMC - PubMed
-
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

