Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly
- PMID: 28760201
- PMCID: PMC5538823
- DOI: 10.7554/eLife.25317
Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly
Erratum in
-
Correction: Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly.Elife. 2017 Sep 6;6:e31641. doi: 10.7554/eLife.31641. Elife. 2017. PMID: 28874256 Free PMC article. No abstract available.
Abstract
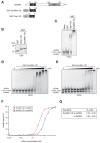
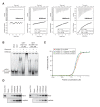
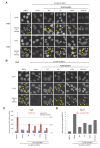
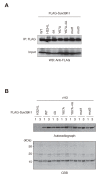
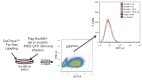
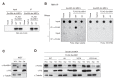
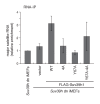
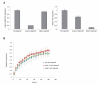
SUV39H is the major histone H3 lysine 9 (H3K9)-specific methyltransferase that targets pericentric regions and is crucial for assembling silent heterochromatin. SUV39H recognizes trimethylated H3K9 (H3K9me3) via its chromodomain (CD), and enriched H3K9me3 allows SUV39H to target specific chromosomal regions. However, the detailed targeting mechanisms, especially for naïve chromatin without preexisting H3K9me3, are poorly understood. Here we show that Suv39h1's CD (Suv39h1-CD) binds nucleic acids, and this binding is important for its function in heterochromatin assembly. Suv39h1-CD had higher binding affinity for RNA than DNA, and its ability to bind nucleic acids was independent of its H3K9me3 recognition. Suv39h1 bound major satellite RNAs in vivo, and knockdown of major satellite RNAs lowered Suv39h1 retention on pericentromere. Suv39h1 mutational studies indicated that both the nucleic acid-binding and H3K9me-binding activities of Suv39h1-CD were crucial for its pericentric heterochromatin assembly. These results suggest that chromatin-bound RNAs contribute to creating SUV39H's target specificity.
Keywords: H3K9 methylation; HP1; Suv39h1; chromodomain; chromosomes; genes; heterochromatin; mouse; pericentromere.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures







Comment in
-
Silencing repetitive DNA.Elife. 2017 Aug 1;6:e29503. doi: 10.7554/eLife.29503. Elife. 2017. PMID: 28760198 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

