Comprehensive Arrayed Transposon Mutant Library of Klebsiella pneumoniae Outbreak Strain KPNIH1
- PMID: 28760848
- PMCID: PMC5637181
- DOI: 10.1128/JB.00352-17
Comprehensive Arrayed Transposon Mutant Library of Klebsiella pneumoniae Outbreak Strain KPNIH1
Abstract
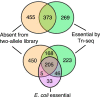
Klebsiella pneumoniae and other carbapenem-resistant members of the family Enterobacteriaceae are a major cause of hospital-acquired infections, yet the basis of their success as nosocomial pathogens is poorly understood. To help provide a foundation for genetic analysis of K. pneumoniae, we created an arrayed, sequence-defined transposon mutant library of an isolate from the 2011 outbreak of infections at the U.S. National Institutes of Health Clinical Center. The library is made up of 12,000 individually arrayed mutants of a carbapenemase deletion parent strain and provides coverage of 85% of the predicted genes. The library includes an average of 2.5 mutants per gene, with most insertion locations identified and confirmed in two independent rounds of Sanger sequencing. On the basis of an independent transposon sequencing assay, about half of the genes lacking representatives in this "two-allele" library are essential for growth on nutrient agar. To validate the use of the library for phenotyping, we screened candidate mutants for increased antibiotic sensitivity by using custom phenotypic microarray plates. This screening identified several mutations increasing sensitivity to β-lactams (in acrB1, mcrB, ompR, phoP1, and slt1) and found that two-component regulator cpxAR mutations increased multiple sensitivities (to an aminoglycoside, a fluoroquinolone, and several β-lactams). Strains making up the two-allele mutant library are available through a web-based request mechanism.IMPORTANCE K. pneumoniae and other carbapenem-resistant members of the family Enterobacteriaceae are recognized as a top public health threat by the Centers for Disease Control and Prevention. The analysis of these major nosocomial pathogens has been limited by the experimental resources available for studying them. The work presented here describes a sequence-defined mutant library of a K. pneumoniae strain (KPNIH1) that represents an attractive model for studies of this pathogen because it is a recent isolate of the major sequence type that causes infection, the epidemiology of the outbreak it caused is well characterized, and an annotated genome sequence is available. The ready availability of defined mutants deficient in nearly all of the nonessential genes of the model strain should facilitate the genetic dissection of complex traits like pathogenesis and antibiotic resistance.
Keywords: Cre; KPC; ST258; cpxR; essential gene; phenotypic microarray.
Copyright © 2017 American Society for Microbiology.
Figures


References
-
- Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi:10.1073/pnas.1321364111. - DOI - PMC - PubMed
-
- Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi:10.1126/scitranslmed.3004129. - DOI - PMC - PubMed
-
- Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, Tzelepi E, Daikos GL. 2013. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother 57:5144–5146. doi:10.1128/AAC.01052-13. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

