Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex
- PMID: 28760978
- PMCID: PMC5565445
- DOI: 10.1073/pnas.1705623114
Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex
Abstract
The synaptonemal complex (SC), a structure highly conserved from yeast to mammals, assembles between homologous chromosomes and is essential for accurate chromosome segregation at the first meiotic division. In Drosophila melanogaster, many SC components and their general positions within the complex have been dissected through a combination of genetic analyses, superresolution microscopy, and electron microscopy. Although these studies provide a 2D understanding of SC structure in Drosophila, the inability to optically resolve the minute distances between proteins in the complex has precluded its 3D characterization. A recently described technology termed expansion microscopy (ExM) uniformly increases the size of a biological sample, thereby circumventing the limits of optical resolution. By adapting the ExM protocol to render it compatible with structured illumination microscopy, we can examine the 3D organization of several known Drosophila SC components. These data provide evidence that two layers of SC are assembled. We further speculate that each SC layer may connect two nonsister chromatids, and present a 3D model of the Drosophila SC based on these findings.
Keywords: expansion microscopy; meiosis; sister chromatids; structured illumination microscopy; synaptonemal complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

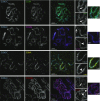
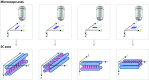


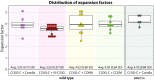
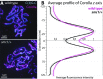
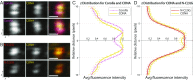
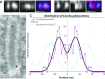
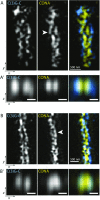

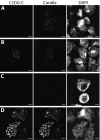
References
-
- Cahoon CK, Hawley RS. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol. 2016;23:369–377. - PubMed
-
- Goldstein P. Multiple synaptonemal complexes (polycomplexes): Origin, structure and function. Cell Biol Int Rep. 1987;11:759–796. - PubMed
-
- Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. - PubMed
-
- Moses MJ, Counce SJ, Paulson DF. Synaptonemal complex complement of man in spreads of spermatocytes, with details of the sex chromosome pair. Science. 1975;187:363–365. - PubMed
-
- Schmekel K, Daneholt B. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 1995;5:239–242. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

