Presumptive TRP channel CED-11 promotes cell volume decrease and facilitates degradation of apoptotic cells in Caenorhabditis elegans
- PMID: 28760991
- PMCID: PMC5565440
- DOI: 10.1073/pnas.1705084114
Presumptive TRP channel CED-11 promotes cell volume decrease and facilitates degradation of apoptotic cells in Caenorhabditis elegans
Abstract
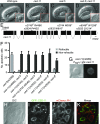
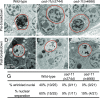
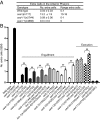
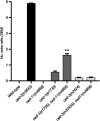
Apoptotic cells undergo a series of morphological changes. These changes are dependent on caspase cleavage of downstream targets, but which targets are significant and how they facilitate the death process are not well understood. In Caenorhabditis elegans an increase in the refractility of the dying cell is a hallmark morphological change that is caspase dependent. We identify a presumptive transient receptor potential (TRP) cation channel, CED-11, that acts in the dying cell to promote the increase in apoptotic cell refractility. CED-11 is required for multiple other morphological changes during apoptosis, including an increase in electron density as visualized by electron microscopy and a decrease in cell volume. In ced-11 mutants, the degradation of apoptotic cells is delayed. Mutation of ced-11 does not cause an increase in cell survival but can enhance cell survival in other cell-death mutants, indicating that ced-11 facilitates the death process. In short, ced-11 acts downstream of caspase activation to promote the shrinkage, death, and degradation of apoptotic cells.
Keywords: C. elegans; TRP channel; apoptosis; apoptotic volume decrease; cell volume.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. - PubMed
-
- Igney FH, Krammer PH. Death and anti-death: Tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–288. - PubMed
-
- Lüthi AU, Martin SJ. The CASBAH: A searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

