Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids
- PMID: 28761003
- PMCID: PMC5565447
- DOI: 10.1073/pnas.1705815114
Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids
Abstract
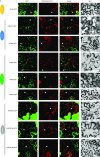
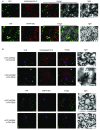
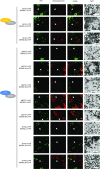
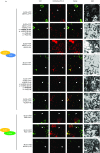
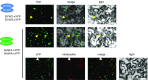
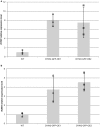
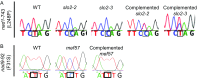
Recent identification of several different types of RNA editing factors in plant organelles suggests complex RNA editosomes within which each factor has a different task. However, the precise protein interactions between the different editing factors are still poorly understood. In this paper, we show that the E+-type pentatricopeptide repeat (PPR) protein SLO2, which lacks a C-terminal cytidine deaminase-like DYW domain, interacts in vivo with the DYW-type PPR protein DYW2 and the P-type PPR protein NUWA in mitochondria, and that the latter enhances the interaction of the former ones. These results may reflect a protein scaffold or complex stabilization role of NUWA between E+-type PPR and DYW2 proteins. Interestingly, DYW2 and NUWA also interact in chloroplasts, and DYW2-GFP overexpressing lines show broad editing defects in both organelles, with predominant specificity for sites edited by E+-type PPR proteins. The latter suggests a coordinated regulation of organellar multiple site editing through DYW2, which probably provides the deaminase activity to E+ editosomes.
Keywords: DYW2; NUWA; PPR; RNA editing; SLO2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













References
-
- Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. - PubMed
-
- Shikanai T. RNA editing in plants: Machinery and flexibility of site recognition. Biochim Biophys Acta. 2015;1847:779–785. - PubMed
-
- Sun T, Bentolila S, Hanson MR. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21:962–973. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

