The Skin Microbiome of Cohabiting Couples
- PMID: 28761935
- PMCID: PMC5527301
- DOI: 10.1128/mSystems.00043-17
The Skin Microbiome of Cohabiting Couples
Abstract
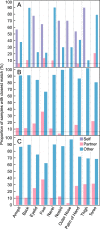
Distinct microbial communities inhabit individuals as part of the human skin microbiome and are continually shed to the surrounding environment. Microbial communities from 17 skin sites of 10 sexually active cohabiting couples (20 individuals) were sampled to test whether cohabitation impacts an individual's skin microbiome, leading to shared skin microbiota among partner pairs. Amplified 16S rRNA genes of bacteria and archaea from a total of 340 skin swabs were analyzed by high-throughput sequencing, and the results demonstrated that cohabitation was significantly associated with microbial community composition, although this association was greatly exceeded by characteristics of body location and individuality. Random forest modeling demonstrated that the partners could be predicted 86% of the time (P < 0.001) based on their skin microbiome profiles, which was always greater than combinations of incorrectly matched partners. Cohabiting couples had the most similar overall microbial skin communities on their feet, according to Bray-Curtis distances. In contrast, thigh microbial communities were strongly associated with biological sex rather than cohabiting partner. Additional factors that were associated with the skin microbiome of specific body locations included the use of skin care products, pet ownership, allergies, and alcohol consumption. These baseline data identified links between the skin microbiome and daily interactions among cohabiting individuals, adding to known factors that shape the human microbiome and, by extension, its relation to human health. IMPORTANCE Our work characterizes the influence of cohabitation as a factor influencing the composition of the skin microbiome. Although the body site and sampled individual were stronger influences than other factors collected as metadata in this study, we show that modeling of detected microbial taxa can help with correct identifications of cohabiting partners based on skin microbiome profiles using machine learning approaches. These results show that a cohabiting partner can significantly influence our microbiota. Follow-up studies will be important for investigating the implications of shared microbiota on dermatological health and the contributions of cohabiting parents to the microbiome profiles of their infants.
Keywords: cohabitation; high-throughput sequencing; human skin; microbiome; random forest modeling.
Figures




References
-
- Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, Berg-Lyon D, Ackermann G, Moeller Christensen GJ, Nakatsuji T, Zhang L, Borkowski AW, Meehan MJ, Dorrestein K, Gallo RL, Bandeira N, Knight R, Alexandrov T, Dorrestein PC. 2015. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci U S A 112:E2120–E2129. doi: 10.1073/pnas.1424409112. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
