Genome-Wide Sensitivity Analysis of the Microsymbiont Sinorhizobium meliloti to Symbiotically Important, Defensin-Like Host Peptides
- PMID: 28765224
- PMCID: PMC5539429
- DOI: 10.1128/mBio.01060-17
Genome-Wide Sensitivity Analysis of the Microsymbiont Sinorhizobium meliloti to Symbiotically Important, Defensin-Like Host Peptides
Abstract
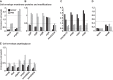
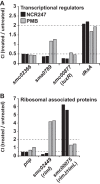
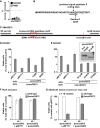
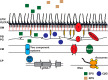
The model legume species Medicago truncatula expresses more than 700 nodule-specific cysteine-rich (NCR) signaling peptides that mediate the differentiation of Sinorhizobium meliloti bacteria into nitrogen-fixing bacteroids. NCR peptides are essential for a successful symbiosis in legume plants of the inverted-repeat-lacking clade (IRLC) and show similarity to mammalian defensins. In addition to signaling functions, many NCR peptides exhibit antimicrobial activity in vitro and in vivo Bacterial resistance to these antimicrobial activities is likely to be important for symbiosis. However, the mechanisms used by S. meliloti to resist antimicrobial activity of plant peptides are poorly understood. To address this, we applied a global genetic approach using transposon mutagenesis followed by high-throughput sequencing (Tn-seq) to identify S. meliloti genes and pathways that increase or decrease bacterial competitiveness during exposure to the well-studied cationic NCR247 peptide and also to the unrelated model antimicrobial peptide polymyxin B. We identified 78 genes and several diverse pathways whose interruption alters S. meliloti resistance to NCR247. These genes encode the following: (i) cell envelope polysaccharide biosynthesis and modification proteins, (ii) inner and outer membrane proteins, (iii) peptidoglycan (PG) effector proteins, and (iv) non-membrane-associated factors such as transcriptional regulators and ribosome-associated factors. We describe a previously uncharacterized yet highly conserved peptidase, which protects S. meliloti from NCR247 and increases competitiveness during symbiosis. Additionally, we highlight a considerable number of uncharacterized genes that provide the basis for future studies to investigate the molecular basis of symbiotic development as well as chronic pathogenic interactions.IMPORTANCE Soil rhizobial bacteria enter into an ecologically and economically important symbiotic interaction with legumes, in which they differentiate into physiologically distinct bacteroids that provide essential ammonia to the plant in return for carbon sources. Plant signal peptides are essential and specific to achieve these physiological changes. These peptides show similarity to mammalian defensin peptides which are part of the first line of defense to control invading bacterial populations. A number of these legume peptides are indeed known to possess antimicrobial activity, and so far, only the bacterial BacA protein is known to protect rhizobial bacteria against their antimicrobial action. This study identified numerous additional bacterial factors that mediate protection and belong to diverse biological pathways. Our results significantly contribute to our understanding of the molecular roles of bacterial factors during legume symbioses and, second, provide insights into the mechanisms that pathogenic bacteria may use to resist the antimicrobial effects of defensins during infections.
Keywords: antimicrobial peptides; host-microbe interactions; symbiosis.
Copyright © 2017 Arnold et al.
Figures


References
-
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132:161–173. doi:10.1104/pp.102.018192. - DOI - PMC - PubMed
-
- Van De Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, Satiat-Jeunemaître B, Alunni B, Bourge M, Kucho KI, Abe M, Kereszt A, Maroti G, Uchiumi T, Kondorosi E, Mergaert P. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126. doi:10.1126/science.1184057. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

