Bacterial membrane vesicles transport their DNA cargo into host cells
- PMID: 28765539
- PMCID: PMC5539193
- DOI: 10.1038/s41598-017-07288-4
Bacterial membrane vesicles transport their DNA cargo into host cells
Abstract
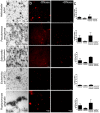
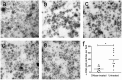
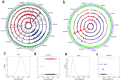
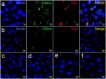
Bacterial outer membrane vesicles (OMVs) are extracellular sacs containing biologically active products, such as proteins, cell wall components and toxins. OMVs are reported to contain DNA, however, little is known about the nature of this DNA, nor whether it can be transported into host cells. Our work demonstrates that chromosomal DNA is packaged into OMVs shed by bacteria during exponential phase. Most of this DNA was present on the external surfaces of OMVs, with smaller amounts located internally. The DNA within the internal compartments of Pseudomonas aeruginosa OMVs were consistently enriched in specific regions of the bacterial chromosome, encoding proteins involved in virulence, stress response, antibiotic resistance and metabolism. Furthermore, we demonstrated that OMVs carry DNA into eukaryotic cells, and this DNA was detectable by PCR in the nuclear fraction of cells. These findings suggest a role for OMV-associated DNA in bacterial-host cell interactions and have implications for OMV-based vaccines.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Lee, E.-Y., Choi, D.-Y., Kim, D.-K. & Kim, J.-W. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics9, 5425–5436 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

