High-Resolution Patterned Cellular Constructs by Droplet-Based 3D Printing
- PMID: 28765636
- PMCID: PMC5539110
- DOI: 10.1038/s41598-017-06358-x
High-Resolution Patterned Cellular Constructs by Droplet-Based 3D Printing
Abstract
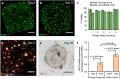
Bioprinting is an emerging technique for the fabrication of living tissues that allows cells to be arranged in predetermined three-dimensional (3D) architectures. However, to date, there are limited examples of bioprinted constructs containing multiple cell types patterned at high-resolution. Here we present a low-cost process that employs 3D printing of aqueous droplets containing mammalian cells to produce robust, patterned constructs in oil, which were reproducibly transferred to culture medium. Human embryonic kidney (HEK) cells and ovine mesenchymal stem cells (oMSCs) were printed at tissue-relevant densities (107 cells mL-1) and a high droplet resolution of 1 nL. High-resolution 3D geometries were printed with features of ≤200 μm; these included an arborised cell junction, a diagonal-plane junction and an osteochondral interface. The printed cells showed high viability (90% on average) and HEK cells within the printed structures were shown to proliferate under culture conditions. Significantly, a five-week tissue engineering study demonstrated that printed oMSCs could be differentiated down the chondrogenic lineage to generate cartilage-like structures containing type II collagen.
Conflict of interest statement
Hagan Bayley is the Founder, a Director and a shareholder of OxSyBio, a company engaged in the development of printed tissues and tissue-like materials. Work in the Bayley laboratory at the University of Oxford is supported in part by OxSyBio. During revision of the paper, Alexander Graham, Sam Olof, Madeline Burke and Stuart Box became employees of OxSyBio.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

