ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference
- PMID: 28767641
- PMCID: PMC5777669
- DOI: 10.1038/nature23671
ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference
Abstract
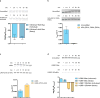
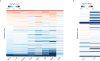
ATP-dependent chromatin remodellers regulate access to genetic information by controlling nucleosome positions in vivo. However, the mechanism by which remodellers discriminate between different nucleosome substrates is poorly understood. Many chromatin remodelling proteins possess conserved protein domains that interact with nucleosomal features. Here we used a quantitative high-throughput approach, based on the use of a DNA-barcoded mononucleosome library, to profile the biochemical activity of human ISWI family remodellers in response to a diverse set of nucleosome modifications. We show that accessory (non-ATPase) subunits of ISWI remodellers can distinguish between differentially modified nucleosomes, directing remodelling activity towards specific nucleosome substrates according to their modification state. Unexpectedly, we show that the nucleosome acidic patch is necessary for maximum activity of all ISWI remodellers evaluated. This dependence also extends to CHD and SWI/SNF family remodellers, suggesting that the acidic patch may be generally required for chromatin remodelling. Critically, remodelling activity can be regulated by modifications neighbouring the acidic patch, signifying that it may act as a tunable interaction hotspot for ATP-dependent chromatin remodellers and, by extension, many other chromatin effectors that engage this region of the nucleosome surface.
Conflict of interest statement
The authors declare no competing financial interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

