Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation
- PMID: 28768204
- PMCID: PMC5554778
- DOI: 10.1016/j.celrep.2017.07.009
Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation
Abstract
The mouse inner cell mass (ICM) segregates into the epiblast and primitive endoderm (PrE) lineages coincident with implantation of the embryo. The epiblast subsequently undergoes considerable expansion of cell numbers prior to gastrulation. To investigate underlying regulatory principles, we performed systematic single-cell RNA sequencing (seq) of conceptuses from E3.5 to E6.5. The epiblast shows reactivation and subsequent inactivation of the X chromosome, with Zfp57 expression associated with reactivation and inactivation together with other candidate regulators. At E6.5, the transition from epiblast to primitive streak is linked with decreased expression of polycomb subunits, suggesting a key regulatory role. Notably, our analyses suggest elevated transcriptional noise at E3.5 and within the non-committed epiblast at E6.5, coinciding with exit from pluripotency. By contrast, E6.5 primitive streak cells became highly synchronized and exhibit a shortened G1 cell-cycle phase, consistent with accelerated proliferation. Our study systematically charts transcriptional noise and uncovers molecular processes associated with early lineage decisions.
Keywords: X-chromosome; embryo; epiblast; gastrulation; primitive endoderm; primitive streak; single-cell RNA-seq; transcriptional noise.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


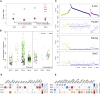
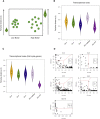


Comment in
-
Cell lineage specification at single cell resolution.Stem Cell Investig. 2017 Sep 19;4:76. doi: 10.21037/sci.2017.09.03. eCollection 2017. Stem Cell Investig. 2017. PMID: 29057248 Free PMC article. No abstract available.
References
-
- Arias A.M., Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 2006;7:34–44. - PubMed
-
- Bar-Even A., Paulsson J., Maheshri N., Carmi M., O’Shea E., Pilpel Y., Barkai N. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–643. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

