Genetic Mapping of Quantitative Trait Loci for Yield-Affecting Traits in a Barley Doubled Haploid Population Derived from Clipper × Sahara 3771
- PMID: 28769936
- PMCID: PMC5513936
- DOI: 10.3389/fpls.2017.00688
Genetic Mapping of Quantitative Trait Loci for Yield-Affecting Traits in a Barley Doubled Haploid Population Derived from Clipper × Sahara 3771
Abstract
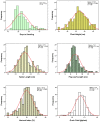
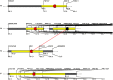
Many traits play essential roles in determining crop yield. Wide variation for morphological traits exists in Hordeum vulgare L., but the genetic basis of this morphological variation is largely unknown. To understand genetic basis controlling morphological traits affecting yield, a barley doubled haploid population (146 individuals) derived from Clipper × Sahara 3771 was used to map chromosome regions underlying days to awn appearance, plant height, fertile spike number, flag leaf length, spike length, harvest index, seed number per plant, thousands kernel weight, and grain yield. Twenty-seven QTLs for nine traits were mapped to the barley genome that described 3-69% of phenotypic variations; and some genomic regions harbor a given QTL for more than one trait. Out of 27 QTLs identified, 19 QTLs were novel. Chromosomal regions on 1H, 2H, 4H, and 6H associated with seed grain yield, and chromosome regions on 2H and 6H had major effects on grain yield (GY). One major QTL for seed number per plant was flanked by marker VRS1-KSUF15 on chromosome 2H. This QTL was also associated with GY. Some loci controlling thousands kernel weight (TKW), fertile spike number (FSN), and GY were the same. The major grain yield QTL detected on linkage PSR167 co-localized with TAM10. Two major QTLs controlling TKW and FSN were also mapped at this locus. Eight QTLs on chromosomes 1H, 2H, 3H, 4H, 5H, 6H, and 7H consistently affected spike characteristics. One major QTL (ANIONT1A-TACMD) on 4H affected both spike length (SL) and spike number explained 9 and 5% of the variation of SL and FSN, respectively. In conclusion, this study could cast some light on the genetic basis of the studied pivotal traits. Moreover, fine mapping of the identified major effect markers may facilitate the application of molecular markers in barley breeding programs.
Keywords: barley (Hordeum vulgare L.); grain yield; linkage analysis; morphological traits; quantitative trait loci (QTL).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aminfar Z., Siahsar B. A., Heidary M. (2011). Determination of chromosomes controlling physiological traits associated to salt tolerance in barley in seedling stage. New. Biotechnol. 25:304. 10.1016/j.nbt.2009.06.762 - DOI
-
- Arpaili D., Yagmur M. (2015). The determination of selection criteria using path analysis in two rowed barley (Hordeum vulgare L. Conv. Distichon). Turk. J. Agric. Nat. Sci. 2 248–255.
-
- Beecher B., Smidansky E. D., See D., Blake T. K., Giroux M. J. (2001). Mapping and sequence analysis of barley hordoindo lines. Theor. Appl. Genet. 102 833–840. 10.1007/s001220000488 - DOI
-
- Bezant J., Laurie D., Pratchett N., Chojeckls J., Kearsey M. (1996). Marker regression mapping of QTL controlling flowering time and plant height in a spring barley (Hordeum vulgare L.) cross. Heredity 77 64–73. 10.1038/hdy.1996.109 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

