Unraveling the Root Proteome Changes and Its Relationship to Molecular Mechanism Underlying Salt Stress Response in Radish (Raphanus sativus L.)
- PMID: 28769938
- PMCID: PMC5509946
- DOI: 10.3389/fpls.2017.01192
Unraveling the Root Proteome Changes and Its Relationship to Molecular Mechanism Underlying Salt Stress Response in Radish (Raphanus sativus L.)
Abstract
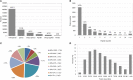
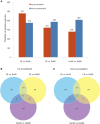
To understand the molecular mechanism underlying salt stress response in radish, iTRAQ-based proteomic analysis was conducted to investigate the differences in protein species abundance under different salt treatments. In total, 851, 706, and 685 differential abundance protein species (DAPS) were identified between CK vs. Na100, CK vs. Na200, and Na100 vs. Na200, respectively. Functional annotation analysis revealed that salt stress elicited complex proteomic alterations in radish roots involved in carbohydrate and energy metabolism, protein metabolism, signal transduction, transcription regulation, stress and defense and transport. Additionally, the expression levels of nine genes encoding DAPS were further verified using RT-qPCR. The integrative analysis of transcriptomic and proteomic data in conjunction with miRNAs was further performed to strengthen the understanding of radish response to salinity. The genes responsible for signal transduction, ROS scavenging and transport activities as well as several key miRNAs including miR171, miR395, and miR398 played crucial roles in salt stress response in radish. Based on these findings, a schematic genetic regulatory network of salt stress response was proposed. This study provided valuable insights into the molecular mechanism underlying salt stress response in radish roots and would facilitate developing effective strategies toward genetically engineered salt-tolerant radish and other root vegetable crops.
Keywords: association analysis; iTRAQ; proteomics; radish; salt stress.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

