The RNA polymerase II CTD "orphan" residues: Emerging insights into the functions of Tyr-1, Thr-4, and Ser-7
- PMID: 28771071
- PMCID: PMC5791814
- DOI: 10.1080/21541264.2017.1338176
The RNA polymerase II CTD "orphan" residues: Emerging insights into the functions of Tyr-1, Thr-4, and Ser-7
Abstract
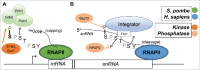
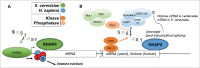
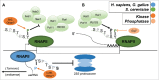
The C-terminal domain (CTD) of the RNA polymerase II largest subunit consists of a unique repeated heptad sequence of the consensus Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. An important function of the CTD is to couple transcription with RNA processing reactions that occur during the initiation, elongation, and termination phases of transcription. During this transcription cycle, the CTD is subject to extensive modification, primarily phosphorylation, on its non-proline residues. Reversible phosphorylation of Ser2 and Ser5 is well known to play important and general functions during transcription in all eukaryotes. More recent studies have enhanced our understanding of Tyr1, Thr4, and Ser7, and what have been previously characterized as unknown or specialized functions for these residues has changed to a more fine-detailed map of transcriptional regulation that highlights similarities as well as significant differences between organisms. Here, we review recent findings on the function and modification of these three residues, which further illustrate the importance of the CTD in precisely modulating gene expression.
Keywords: RNA processing; kinases; phosphatases; phosphorylation; transcription.
Figures
References
-
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012; 26:2119–2137; PMID:23028141; https://doi.org/10.1101/gad.200303.112 - DOI - PMC - PubMed
-
- Corden JL. RNA polymerase II C-Terminal domain: tethering transcription to transcript and template. Chem Rev 2013; 113:8423–8455; PMID:24040939; https://doi.org/10.1021/cr400158h - DOI - PMC - PubMed
-
- Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 2013; 1829:55–62; PMID:22982363; https://doi.org/10.1016/j.bbagrm.2012.08.013 - DOI - PubMed
-
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 2014; 15:163–175; PMID:24514444; https://doi.org/10.1038/nrg3662 - DOI - PMC - PubMed
-
- Conaway RC, Conaway JW. Orchestrating transcription with the pol II CTD. Nat Rev Mol Cell Biol 2015; 16:128; PMID:25693123; https://doi.org/10.1038/nrm3956 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
