Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes
- PMID: 28771184
- PMCID: PMC5578081
- DOI: 10.3390/ijms18081691
Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes
Abstract
Bacterial photoreceptors absorb light energy and transform it into intracellular signals that regulate metabolism. Bacterial phytochrome photoreceptors (BphPs), some cyanobacteriochromes (CBCRs) and allophycocyanins (APCs) possess the near-infrared (NIR) absorbance spectra that make them promising molecular templates to design NIR fluorescent proteins (FPs) and biosensors for studies in mammalian cells and whole animals. Here, we review structures, photochemical properties and molecular functions of several families of bacterial photoreceptors. We next analyze molecular evolution approaches to develop NIR FPs and biosensors. We then discuss phenotypes of current BphP-based NIR FPs and compare them with FPs derived from CBCRs and APCs. Lastly, we overview imaging applications of NIR FPs in live cells and in vivo. Our review provides guidelines for selection of existing NIR FPs, as well as engineering approaches to develop NIR FPs from the novel natural templates such as CBCRs.
Keywords: allophycocyanin; bacterial photoreceptor; cyanobacteriochrome; near-infrared fluorescent protein; phytochrome; tetrapyrrole.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
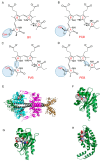
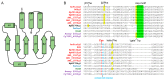

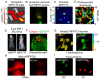
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

