Structural insight into the activation of a class B G-protein-coupled receptor by peptide hormones in live human cells
- PMID: 28771403
- PMCID: PMC5542768
- DOI: 10.7554/eLife.27711
Structural insight into the activation of a class B G-protein-coupled receptor by peptide hormones in live human cells
Abstract
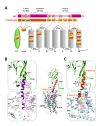
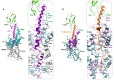
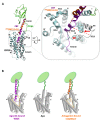
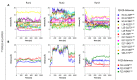
The activation mechanism of class B G-protein-coupled receptors (GPCRs) remains largely unknown. To characterize conformational changes induced by peptide hormones, we investigated interactions of the class B corticotropin-releasing factor receptor type 1 (CRF1R) with two peptide agonists and three peptide antagonists obtained by N-truncation of the agonists. Surface mapping with genetically encoded photo-crosslinkers and pair-wise crosslinking revealed distinct footprints of agonists and antagonists on the transmembrane domain (TMD) of CRF1R and identified numerous ligand-receptor contact sites, directly from the intact receptor in live human cells. The data enabled generating atomistic models of CRF- and CRF(12-41)-bound CRF1R, further explored by molecular dynamics simulations. We show that bound agonist and antagonist adopt different folds and stabilize distinct TMD conformations, which involves bending of helices VI and VII around flexible glycine hinges. Conservation of these glycine hinges among all class B GPCRs suggests their general role in activation of these receptors.
Keywords: biochemistry; bioorthogonal crosslinking; biophysics; class B GPCRs; expanded genetic code; human; molecular modeling; orthosteric antagonism; peptide ligands; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
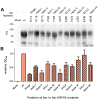

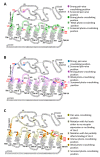


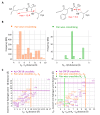




Similar articles
-
Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR.Cell. 2013 Dec 5;155(6):1258-69. doi: 10.1016/j.cell.2013.11.008. Epub 2013 Nov 27. Cell. 2013. PMID: 24290358 Free PMC article.
-
Understanding Corticotropin Releasing Factor Receptor (CRFR) Activation Using Structural Models.Curr Mol Pharmacol. 2017;10(4):325-333. doi: 10.2174/1874467210666170110122939. Curr Mol Pharmacol. 2017. PMID: 28103783 Review.
-
Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.Curr Mol Pharmacol. 2017;10(4):334-344. doi: 10.2174/1874467210666170110114727. Curr Mol Pharmacol. 2017. PMID: 28183242 Free PMC article.
-
Structural-functional analysis of the third transmembrane domain of the corticotropin-releasing factor type 1 receptor: role in activation and allosteric antagonism.J Biol Chem. 2014 Jul 4;289(27):18966-77. doi: 10.1074/jbc.M113.544460. Epub 2014 May 16. J Biol Chem. 2014. PMID: 24838244 Free PMC article.
-
Structures of the First Extracellular Domain of CRF Receptors.Curr Mol Pharmacol. 2017;10(4):318-324. doi: 10.2174/1874467210666170110120301. Curr Mol Pharmacol. 2017. PMID: 28103782 Review.
Cited by
-
Crosslinking-guided geometry of a complete CXC receptor-chemokine complex and the basis of chemokine subfamily selectivity.PLoS Biol. 2020 Apr 9;18(4):e3000656. doi: 10.1371/journal.pbio.3000656. eCollection 2020 Apr. PLoS Biol. 2020. PMID: 32271748 Free PMC article.
-
Non-canonical amino acid incorporation into AAV5 capsid enhances lung transduction in mice.Mol Ther Methods Clin Dev. 2023 Oct 7;31:101129. doi: 10.1016/j.omtm.2023.101129. eCollection 2023 Dec 14. Mol Ther Methods Clin Dev. 2023. PMID: 37886602 Free PMC article.
-
Site-Specific Crosslinking Reveals Phosphofructokinase-L Inhibition Drives Self-Assembly and Attenuation of Protein Interactions.bioRxiv [Preprint]. 2023 Sep 20:2023.09.19.558525. doi: 10.1101/2023.09.19.558525. bioRxiv. 2023. Update in: Adv Biol Regul. 2023 Dec;90:100987. doi: 10.1016/j.jbior.2023.100987. PMID: 37781627 Free PMC article. Updated. Preprint.
-
New Insights Into the Evolution of Corticotropin-Releasing Hormone Family With a Special Focus on Teleosts.Front Endocrinol (Lausanne). 2022 Jul 22;13:937218. doi: 10.3389/fendo.2022.937218. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35937826 Free PMC article.
-
Single-Molecule Imaging of Integral Membrane Protein Dynamics and Function.Annu Rev Biophys. 2024 Jul;53(1):427-453. doi: 10.1146/annurev-biophys-070323-024308. Annu Rev Biophys. 2024. PMID: 39013028 Free PMC article. Review.
References
-
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. Journal of Chemical Theory and Computation. 2012;8:3257–3273. doi: 10.1021/ct300400x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

