Additional value of screening for minor genes and copy number variants in hypertrophic cardiomyopathy
- PMID: 28771489
- PMCID: PMC5542623
- DOI: 10.1371/journal.pone.0181465
Additional value of screening for minor genes and copy number variants in hypertrophic cardiomyopathy
Abstract
Introduction: Hypertrophic cardiomyopathy (HCM) is the most prevalent inherited heart disease. Next-generation sequencing (NGS) is the preferred genetic test, but the diagnostic value of screening for minor and candidate genes, and the role of copy number variants (CNVs) deserves further evaluation.
Methods: Three hundred and eighty-seven consecutive unrelated patients with HCM were screened for genetic variants in the 5 most frequent genes (MYBPC3, MYH7, TNNT2, TNNI3 and TPM1) using Sanger sequencing (N = 84) or NGS (N = 303). In the NGS cohort we analyzed 20 additional minor or candidate genes, and applied a proprietary bioinformatics algorithm for detecting CNVs. Additionally, the rate and classification of TTN variants in HCM were compared with 427 patients without structural heart disease.
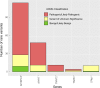
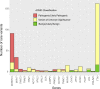
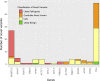
Results: The percentage of patients with pathogenic/likely pathogenic (P/LP) variants in the main genes was 33.3%, without significant differences between the Sanger sequencing and NGS cohorts. The screening for 20 additional genes revealed LP variants in ACTC1, MYL2, MYL3, TNNC1, GLA and PRKAG2 in 12 patients. This approach resulted in more inconclusive tests (36.0% vs. 9.6%, p<0.001), mostly due to variants of unknown significance (VUS) in TTN. The detection rate of rare variants in TTN was not significantly different to that found in the group of patients without structural heart disease. In the NGS cohort, 4 patients (1.3%) had pathogenic CNVs: 2 deletions in MYBPC3 and 2 deletions involving the complete coding region of PLN.
Conclusions: A small percentage of HCM cases without point mutations in the 5 main genes are explained by P/LP variants in minor or candidate genes and CNVs. Screening for variants in TTN in HCM patients drastically increases the number of inconclusive tests, and shows a rate of VUS that is similar to patients without structural heart disease, suggesting that this gene should not be analyzed for clinical purposes in HCM.
Conflict of interest statement
Figures


References
-
- Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–9. - PubMed
-
- Cobo-Marcos M, Cuenca S, Gamez Martinez JM, Bornstein B, Ripoll Vera T, Garcia-Pavia P. Usefulness of genetic testing for hypertrophic cardiomyopathy in real-world practice. Rev Esp Cardiol (Engl Ed). 2013;66(9):746–7. - PubMed
-
- Das KJ, Ingles J, Bagnall RD, Semsarian C. Determining pathogenicity of genetic variants in hypertrophic cardiomyopathy: importance of periodic reassessment. Genet Med. 2014;16(4):286–93. doi: 10.1038/gim.2013.138 - DOI - PubMed
-
- Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880–8. doi: 10.1038/gim.2014.205 - DOI - PubMed
-
- Authors/Task Force m, Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35(39):2733–79. doi: 10.1093/eurheartj/ehu284 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

