Characterization of the genome of a phylogenetically distinct tospovirus and its interactions with the local lesion-induced host Chenopodium quinoa by whole-transcriptome analyses
- PMID: 28771638
- PMCID: PMC5542687
- DOI: 10.1371/journal.pone.0182425
Characterization of the genome of a phylogenetically distinct tospovirus and its interactions with the local lesion-induced host Chenopodium quinoa by whole-transcriptome analyses
Abstract
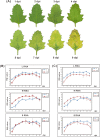
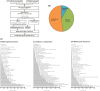
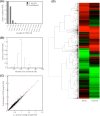
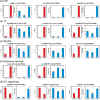
Chenopodium quinoa is a natural local lesion host of numerous plant viruses, including tospoviruses (family Bunyaviridae). Groundnut chlorotic fan-spot tospovirus (GCFSV) has been shown to consistently induce local lesions on the leaves of C. quinoa 4 days post-inoculation (dpi). To reveal the whole genome of GCFSV and its interactions with C. quinoa, RNA-seq was performed to determine the transcriptome profiles of C. quinoa leaves. The high-throughput reads from infected C. quinoa leaves were used to identify the whole genome sequence of GCFSV and its single nucleotide polymorphisms. Our results indicated that GCFSV is a phylogenetically distinct tospovirus. Moreover, 27,170 coding and 29,563 non-coding sequences of C. quinoa were identified through de novo assembly, mixing reads from mock and infected samples. Several key genes involved in the modulation of hypersensitive response (HR) were identified. The expression levels of 4,893 deduced complete genes annotated using the Arabidopsis genome indicated that several HR-related orthologues of pathogenesis-related proteins, transcription factors, mitogen-activated protein kinases, and defense proteins were significantly expressed in leaves that formed local lesions. Here, we also provide new insights into the replication progression of a tospovirus and the molecular regulation of the C. quinoa response to virus infection.
Conflict of interest statement
Figures


References
-
- Pappu HR, Jones RA, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: Successes gained and challenges that lie ahead. Virus Res. 2009; 141:219–236. doi: 10.1016/j.virusres.2009.01.009 - DOI - PubMed
-
- Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A, et al. Bunyaviridae In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus taxonomy: 9th report of the international committee on taxonomy of viruses. New York: Elsevier; 2011. pp. 725–740.
-
- de Haan P, Kormelink R, Resende RO, van Poelwijk F, Peters D, Goldbach R. Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol. 1991; 72:2207–2216. doi: 10.1099/0022-1317-72-9-2207 - DOI - PubMed
-
- van Knippenberg I, Goldbach R, Kormelink R. Purified Tomato spotted wilt virus particles support both genome replication and transcription in vitro. Virology. 2002; 303:278–286. - PubMed
-
- Kormelink R, Storms M, Van Lent J, Peters D, Goldbach R. Expression and subcellular location of the NSm protein of Tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology. 1994; 200:56–65. doi: 10.1006/viro.1994.1162 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

