Rapid Assay to Assess Bacterial Adhesion on Textiles
- PMID: 28773373
- PMCID: PMC5502901
- DOI: 10.3390/ma9040249
Rapid Assay to Assess Bacterial Adhesion on Textiles
Abstract
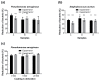
Textiles are frequently colonized by microorganisms leading to undesired consequences like hygienic problems. Biocidal coatings often raise environmental and health concerns, thus sustainable, biocide-free coatings are of interest. To develop novel anti-adhesive textile coatings, a rapid, reliable, and quantitative high-throughput method to study microbial attachment to fabrics is required, however currently not available. Here, a fast and reliable 96-well plate-based screening method is developed. The quantification of bacterial adhesion is based on nucleic acid staining by SYTO9, with Pseudomonas aeruginosa and Staphylococcus aureus as the model microorganisms. Subsequently, 38 commercially available and novel coatings were evaluated for their anti-bacterial adhesion properties. A poly(l-lysine)-g-poly(ethylene glycol) coating on polyester textile substratum revealed an 80% reduction of bacterial adhesion. Both the coating itself and the anti-adhesive property were stable after 20 washing cycles, confirmed by X-ray analysis. The assay provides an efficient tool to rapidly screen for non-biocidal coatings reducing bacterial attachment.
Keywords: antifouling; bacterial adhesion; biofilm; microtiter plate; textile coating.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Szostak-Kotowa J. Biodeterioration of textiles. Int. Biodeterior. Biodegrad. 2004;53:165–170. doi: 10.1016/S0964-8305(03)00090-8. - DOI
-
- Bajpai V., Bajpai S., Jha M.K., Dey A., Ghosh S. Microbial adherence on textile materials: A review. J. Environ. Res. Dev. 2011;5:666–672.
-
- Gao Y., Cranston R. Recent advances in antimicrobial treatments of textiles. Text. Res. J. 2008;78:60–72.
-
- Gouveia I.C. Nanobiotechnology: A new strategy to develop non-toxic antimicrobial textiles for healthcare applications. J. Biotechnol. 2010;150:349. doi: 10.1016/j.jbiotec.2010.09.387. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

