Hiding in Plain Sight: Mining Bacterial Species Records for Phenotypic Trait Information
- PMID: 28776041
- PMCID: PMC5541158
- DOI: 10.1128/mSphere.00237-17
Hiding in Plain Sight: Mining Bacterial Species Records for Phenotypic Trait Information
Abstract
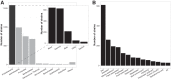
Cultivation in the laboratory is essential for understanding the phenotypic characteristics and environmental preferences of bacteria. However, basic phenotypic information is not readily accessible. Here, we compiled phenotypic and environmental tolerance information for >5,000 bacterial strains described in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) with all information made publicly available in an updatable database. Although the data span 23 different bacterial phyla, most entries described aerobic, mesophilic, neutrophilic strains from Proteobacteria (mainly Alpha- and Gammaproteobacteria), Actinobacteria, Firmicutes, and Bacteroidetes isolated from soils, marine habitats, and plants. Most of the routinely measured traits tended to show a significant phylogenetic signal, although this signal was weak for environmental preferences. We demonstrated how this database could be used to link genomic attributes to differences in pH and salinity optima. We found that adaptations to high salinity or high-pH conditions are related to cell surface transporter genes, along with previously uncharacterized genes that might play a role in regulating environmental tolerances. Together, this work highlights the utility of this database for associating bacterial taxonomy, phylogeny, or specific genes to measured phenotypic traits and emphasizes the need for more comprehensive and consistent measurements of traits across a broader diversity of bacteria. IMPORTANCE Cultivation in the laboratory is key for understanding the phenotypic characteristics, growth requirements, metabolism, and environmental preferences of bacteria. However, oftentimes, phenotypic information is not easily accessible. Here, we compiled phenotypic and environmental tolerance information for >5,000 bacterial strains described in the International Journal of Systematic and Evolutionary Microbiology (IJSEM). We demonstrate how this database can be used to link bacterial taxonomy, phylogeny, or specific genes to measured phenotypic traits and environmental preferences. The phenotypic database can be freely accessed (https://doi.org/10.6084/m9.figshare.4272392), and we have included instructions for researchers interested in adding new entries or curating existing ones.
Keywords: pH; phenotypes; phylogeny; salinity; traits.
Figures

References
-
- Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT, Amann R, Beck BJ, Chain PSG, Chun J, Colwell RR, Danchin A, Dawyndt P, Dedeurwaerdere T, DeLong EF, Detter JC, De Vos P, Donohue TJ, Dong XZ, Ehrlich DS, Fraser C, Gibbs R, Gilbert J, Gilna P, Glöckner FO, Jansson JK, Keasling JD, Knight R, Labeda D, Lapidus A, Lee JS, Li WJ, Ma J, Markowitz V, Moore ERB, Morrison M, Meyer F, Nelson KE, Ohkuma M, Ouzounis CA, Pace N, Parkhill J, Qin N, Rossello-Mora R, Sikorski J, Smith D, Sogin M, Stevens R, Stingl U, Suzuki K-I, Taylor D, Tiedje JM, Tindall B, Wagner M, Weinstock G, Weissenbach J, White O, Wang J, Zhang L, Zhou Y-G, Field D, Whitman WB, Garrity GM, Klenk H-P. 2014. Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol 12:e1001920. doi: 10.1371/journal.pbio.1001920. - DOI - PMC - PubMed
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
