Lignocellulose-Degrading Microbial Communities in Landfill Sites Represent a Repository of Unexplored Biomass-Degrading Diversity
- PMID: 28776044
- PMCID: PMC5541161
- DOI: 10.1128/mSphere.00300-17
Lignocellulose-Degrading Microbial Communities in Landfill Sites Represent a Repository of Unexplored Biomass-Degrading Diversity
Abstract
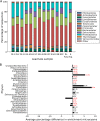
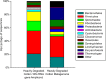
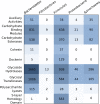
The microbial conversion of lignocellulosic biomass for biofuel production represents a renewable alternative to fossil fuels. However, the discovery of new microbial enzymes with high activity is critical for improving biomass conversion processes. While attempts to identify superior lignocellulose-degrading enzymes have focused predominantly on the animal gut, biomass-degrading communities in landfill sites represent an unexplored resource of hydrolytic enzymes for biomass conversion. Here, to address the paucity of information on biomass-degrading microbial diversity beyond the gastrointestinal tract, cellulose (cotton) "baits" were incubated in landfill leachate microcosms to enrich the landfill cellulolytic microbial community for taxonomic and functional characterization. Metagenome and 16S rRNA gene amplicon sequencing demonstrated the dominance of Firmicutes, Bacteroidetes, Spirochaetes, and Fibrobacteres in the landfill cellulolytic community. Functional metagenome analysis revealed 8,371 carbohydrate active enzymes (CAZymes) belonging to 244 CAZyme families. In addition to observing biomass-degrading enzymes of anaerobic bacterial "cellulosome" systems of members of the Firmicutes, we report the first detection of the Fibrobacter cellulase system and the Bacteroidetes polysaccharide utilization locus (PUL) in landfill sites. These data provide evidence for the presence of multiple mechanisms of biomass degradation in the landfill microbiome and highlight the extraordinary functional diversity of landfill microorganisms as a rich source of biomass-degrading enzymes of potential biotechnological significance. IMPORTANCE The microbial conversion of lignocellulosic biomass for biofuel production represents a renewable alternative to fossil fuels. However, the discovery of new microbial enzymes with high activity is critical for improving biomass conversion processes. While attempts to identify superior lignocellulose-degrading enzymes have focused predominantly on the animal gut, biomass-degrading communities in landfill sites represent an unexplored resource of hydrolytic enzymes for biomass conversion. Here, we identified Firmicutes, Spirochaetes, and Fibrobacteres as key phyla in the landfill cellulolytic community, detecting 8,371 carbohydrate active enzymes (CAZymes) that represent at least three of the recognized strategies for cellulose decomposition. These data highlight substantial hydrolytic enzyme diversity in landfill sites as a source of new enzymes for biomass conversion.
Keywords: Bacteroidetes; CAZymes; Fibrobacter; Firmicutes; Spirochaetes; biomass; cellulose degradation; cultivation; genomics; landfill; metagenomics; microbial ecology.
Figures
References
-
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. - DOI - PubMed
-
- Ilmberger N, Güllert S, Dannenberg J, Rabausch U, Torres J, Wemheuer B, Alawi M, Poehlein A, Chow J, Turaev D, Rattei T, Schmeisser C, Salomon J, Olsen PB, Daniel R, Grundhoff A, Borchert MS, Streit WR. 2014. A comparative metagenome survey of the fecal microbiota of a breast- and a plant-fed Asian elephant reveals an unexpectedly high diversity of glycoside hydrolase family enzymes. PLoS One 9:e106707. doi: 10.1371/journal.pone.0106707. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

