Comprehensive and Automated Linear Interaction Energy Based Binding-Affinity Prediction for Multifarious Cytochrome P450 Aromatase Inhibitors
- PMID: 28776988
- PMCID: PMC5615371
- DOI: 10.1021/acs.jcim.7b00222
Comprehensive and Automated Linear Interaction Energy Based Binding-Affinity Prediction for Multifarious Cytochrome P450 Aromatase Inhibitors
Abstract
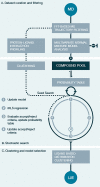
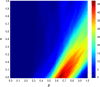
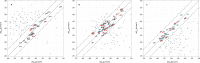
Cytochrome P450 aromatase (CYP19A1) plays a key role in the development of estrogen dependent breast cancer, and aromatase inhibitors have been at the front line of treatment for the past three decades. The development of potent, selective and safer inhibitors is ongoing with in silico screening methods playing a more prominent role in the search for promising lead compounds in bioactivity-relevant chemical space. Here we present a set of comprehensive binding affinity prediction models for CYP19A1 using our automated Linear Interaction Energy (LIE) based workflow on a set of 132 putative and structurally diverse aromatase inhibitors obtained from a typical industrial screening study. We extended the workflow with machine learning methods to automatically cluster training and test compounds in order to maximize the number of explained compounds in one or more predictive LIE models. The method uses protein-ligand interaction profiles obtained from Molecular Dynamics (MD) trajectories to help model search and define the applicability domain of the resolved models. Our method was successful in accounting for 86% of the data set in 3 robust models that show high correlation between calculated and observed values for ligand-binding free energies (RMSE < 2.5 kJ mol-1), with good cross-validation statistics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Integrated Virtual Screening Approach Identifies New CYP19A1 Inhibitors.J Chem Inf Model. 2025 Apr 14;65(7):3529-3543. doi: 10.1021/acs.jcim.5c00204. Epub 2025 Mar 19. J Chem Inf Model. 2025. PMID: 40106333 Free PMC article.
-
Molecular simulations of aromatase reveal new insights into the mechanism of ligand binding.J Chem Inf Model. 2013 Aug 26;53(8):2047-56. doi: 10.1021/ci400225w. Epub 2013 Aug 9. J Chem Inf Model. 2013. PMID: 23927370 Free PMC article.
-
Molecular docking and QSAR study on steroidal compounds as aromatase inhibitors.Eur J Med Chem. 2010 Dec;45(12):5612-20. doi: 10.1016/j.ejmech.2010.09.011. Epub 2010 Sep 17. Eur J Med Chem. 2010. PMID: 20926163
-
Enzymatic and Inhibition Mechanism of Human Aromatase (CYP19A1) Enzyme. A Computational Perspective from QM/MM and Classical Molecular Dynamics Simulations.Mini Rev Med Chem. 2016;16(14):1112-24. doi: 10.2174/1389557516666160623101129. Mini Rev Med Chem. 2016. PMID: 27337972 Review.
-
A review on diverse heterocyclic compounds as the privileged scaffolds in non-steroidal aromatase inhibitors.Bioorg Chem. 2021 Aug;113:105017. doi: 10.1016/j.bioorg.2021.105017. Epub 2021 May 24. Bioorg Chem. 2021. PMID: 34091288 Review.
Cited by
-
Recent Developments in Linear Interaction Energy Based Binding Free Energy Calculations.Front Mol Biosci. 2020 Jun 17;7:114. doi: 10.3389/fmolb.2020.00114. eCollection 2020. Front Mol Biosci. 2020. PMID: 32626725 Free PMC article.
-
The Interaction Mechanism Between C14-Polyacetylene Compounds and the Rat TRPA1 Receptor: An In Silico Study.Int J Mol Sci. 2024 Oct 20;25(20):11290. doi: 10.3390/ijms252011290. Int J Mol Sci. 2024. PMID: 39457072 Free PMC article.
-
Docking Studies and Molecular Dynamics Simulation of Ipomoea batatas L. Leaves Compounds as Lipoxygenase (LOX) Inhibitor.J Pharm Bioallied Sci. 2020 Nov;12(Suppl 2):S836-S840. doi: 10.4103/jpbs.JPBS_103_20. Epub 2020 Nov 5. J Pharm Bioallied Sci. 2020. PMID: 33828386 Free PMC article.
-
A Comparative Linear Interaction Energy and MM/PBSA Study on SIRT1-Ligand Binding Free Energy Calculation.J Chem Inf Model. 2019 Sep 23;59(9):4018-4033. doi: 10.1021/acs.jcim.9b00609. Epub 2019 Sep 11. J Chem Inf Model. 2019. PMID: 31461271 Free PMC article.
-
How Good is Jarzynski's Equality for Computer-Aided Drug Design?J Phys Chem B. 2020 Jul 2;124(26):5338-5349. doi: 10.1021/acs.jpcb.0c02009. Epub 2020 Jun 22. J Phys Chem B. 2020. PMID: 32484689 Free PMC article.
References
-
- Kellis J. T.; Vickery L. E. Purification and characterization of human placental aromatase cytochrome P-450. J. Biol. Chem. 1987, 262, 4413–4420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

