Targeting RNA-Polymerase I in Both Chemosensitive and Chemoresistant Populations in Epithelial Ovarian Cancer
- PMID: 28778862
- PMCID: PMC5668148
- DOI: 10.1158/1078-0432.CCR-17-0282
Targeting RNA-Polymerase I in Both Chemosensitive and Chemoresistant Populations in Epithelial Ovarian Cancer
Abstract
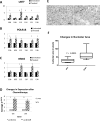
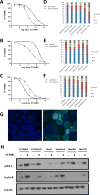
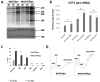
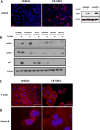
Purpose: A hallmark of neoplasia is increased ribosome biogenesis, and targeting this process with RNA polymerase I (Pol I) inhibitors has shown some efficacy. We examined the contribution and potential targeting of ribosomal machinery in chemotherapy-resistant and -sensitive models of ovarian cancer.Experimental Design: Pol I machinery expression was examined, and subsequently targeted with the Pol I inhibitor CX-5461, in ovarian cancer cell lines, an immortalized surface epithelial line, and patient-derived xenograft (PDX) models with and without chemotherapy. Effects on viability, Pol I occupancy of rDNA, ribosomal content, and chemosensitivity were examined.Results: In PDX models, ribosomal machinery components were increased in chemotherapy-treated tumors compared with controls. Thirteen cell lines were sensitive to CX-5461, with IC50s 25 nmol/L-2 μmol/L. Interestingly, two chemoresistant lines were 10.5- and 5.5-fold more sensitive than parental lines. CX-5461 induced DNA damage checkpoint activation and G2-M arrest with increased γH2AX staining. Chemoresistant cells had 2- to 4-fold increased rDNA Pol I occupancy and increased rRNA synthesis, despite having slower proliferation rates, whereas ribosome abundance and translational efficiency were not impaired. In five PDX models treated with CX-5461, one showed a complete response, one a 55% reduction in tumor volume, and one maintained stable disease for 45 days.Conclusions: Pol I inhibition with CX-5461 shows high activity in ovarian cancer cell lines and PDX models, with an enhanced effect on chemoresistant cells. Effects occur independent of proliferation rates or dormancy. This represents a novel therapeutic approach that may have preferential activity in chemoresistant populations. Clin Cancer Res; 23(21); 6529-40. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
References
-
- Berek JS, Crum C, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2012;119(Suppl):S118–29. - PubMed
-
- Armstrong DK. Relapsed ovarian cancer: challenges and management strategies for a chronic disease. Oncologist. 2002;7(Suppl 5):20–8. - PubMed
-
- Rubin SC, Randall TC, Armstrong KA, et al. Ten-year follow-up of ovarian cancer patients after second-look laparotomy with negative findings. Obstet Gynecol. 1999;93:21–4. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

