Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation
- PMID: 28778989
- PMCID: PMC5584112
- DOI: 10.1084/jem.20160999
Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation
Abstract
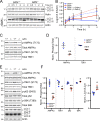
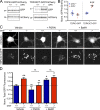
Loss-of-function mutations in GRN cause frontotemporal dementia (FTD) with transactive response DNA-binding protein of 43 kD (TDP-43)-positive inclusions and neuronal ceroid lipofuscinosis (NCL). There are no disease-modifying therapies for either FTD or NCL, in part because of a poor understanding of how mutations in genes such as GRN contribute to disease pathogenesis and neurodegeneration. By studying mice lacking progranulin (PGRN), the protein encoded by GRN, we discovered multiple lines of evidence that PGRN deficiency results in impairment of autophagy, a key cellular degradation pathway. PGRN-deficient mice are sensitive to Listeria monocytogenes because of deficits in xenophagy, a specialized form of autophagy that mediates clearance of intracellular pathogens. Cells lacking PGRN display reduced autophagic flux, and pathological forms of TDP-43 typically cleared by autophagy accumulate more rapidly in PGRN-deficient neurons. Our findings implicate autophagy as a novel therapeutic target for GRN-associated NCL and FTD and highlight the emerging theme of defective autophagy in the broader FTD/amyotrophic lateral sclerosis spectrum of neurodegenerative disease.
© 2017 Chang et al.
Figures




References
-
- Ahmed Z., Sheng H., Xu Y.-F., Lin W.-L., Innes A.E., Gass J., Yu X., Wuertzer C.A., Hou H., Chiba S., et al. . 2010. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 177:311–324. 10.2353/ajpath.2010.090915 - DOI - PMC - PubMed
-
- Almeida M.R., Macário M.C., Ramos L., Baldeiras I., Ribeiro M.H., and Santana I.. 2016. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging. 41:200.e1–200.e5. 10.1016/j.neurobiolaging.2016.02.019 - DOI - PubMed
-
- Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. . 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 442:916–919. 10.1038/nature05016 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

