Rare, protein-truncating variants in ATM, CHEK2 and PALB2, but not XRCC2, are associated with increased breast cancer risks
- PMID: 28779002
- PMCID: PMC5740532
- DOI: 10.1136/jmedgenet-2017-104588
Rare, protein-truncating variants in ATM, CHEK2 and PALB2, but not XRCC2, are associated with increased breast cancer risks
Abstract
Background: Breast cancer (BC) is the most common malignancy in women and has a major heritable component. The risks associated with most rare susceptibility variants are not well estimated. To better characterise the contribution of variants in ATM, CHEK2, PALB2 and XRCC2, we sequenced their coding regions in 13 087 BC cases and 5488 controls from East Anglia, UK.
Methods: Gene coding regions were enriched via PCR, sequenced, variant called and filtered for quality. ORs for BC risk were estimated separately for carriers of truncating variants and of rare missense variants, which were further subdivided by functional domain and pathogenicity as predicted by four in silico algorithms.
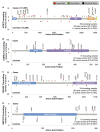
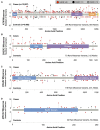
Results: Truncating variants in PALB2 (OR=4.69, 95% CI 2.27 to 9.68), ATM (OR=3.26; 95% CI 1.82 to 6.46) and CHEK2 (OR=3.11; 95% CI 2.15 to 4.69), but not XRCC2 (OR=0.94; 95% CI 0.26 to 4.19) were associated with increased BC risk. Truncating variants in ATM and CHEK2 were more strongly associated with risk of oestrogen receptor (ER)-positive than ER-negative disease, while those in PALB2 were associated with similar risks for both subtypes. There was also some evidence that missense variants in ATM, CHEK2 and PALB2 may contribute to BC risk, but larger studies are necessary to quantify the magnitude of this effect.
Conclusions: Truncating variants in PALB2 are associated with a higher risk of BC than those in ATM or CHEK2. A substantial risk of BC due to truncating XRCC2 variants can be excluded.
Keywords: Cancer: breast; Evidence Based Practice; Genetic Epidemiology; Geneticscreening/counselling.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2017. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures

References
-
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78–85. 10.1056/NEJM200007133430201 - DOI - PubMed
-
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Müller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guénel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Müller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labrèche F, Dumont M, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Brüning T, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dörk T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF; Breast and Ovarian Cancer Susceptibility Collaboration Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) kConFab Investigators Australian Ovarian Cancer Study Group GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network. Large-scale genotyping identifies 41 new loci associated with breast Cancer risk. Nat Genet 2013;45:353–61. 10.1038/ng.2563 - DOI - PMC - PubMed
-
- Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, Devilee P, Meindl A, Couch FJ, Southey M, Goldgar DE, Evans DG, Chenevix-Trench G, Rahman N, Robson M, Domchek SM, Foulkes WD. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015;372:2243–57. 10.1056/NEJMsr1501341 - DOI - PMC - PubMed
-
- Tavtigian SV, Oefner PJ, Babikyan D, Hartmann A, Healey S, Le Calvez-Kelm F, Lesueur F, Byrnes GB, Chuang SC, Forey N, Feuchtinger C, Gioia L, Hall J, Hashibe M, Herte B, McKay-Chopin S, Thomas A, Vallée MP, Voegele C, Webb PM, Whiteman DC, Sangrajrang S, Hopper JL, Southey MC, Andrulis IL, John EM, Chenevix-Trench G, Rare C-TG. Australian Cancer StudyBreast Cancer Family Registries (BCFR)Kathleen Cuningham Foundation Consortium for Research into Familial Aspects of Breast Cancer (kConFab). Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast Cancer. Am J Hum Genet 2009;85:427–46. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
