cAMP receptor protein regulates mouse colonization, motility, fimbria-mediated adhesion, and stress tolerance in uropathogenic Proteus mirabilis
- PMID: 28779108
- PMCID: PMC5544767
- DOI: 10.1038/s41598-017-07304-7
cAMP receptor protein regulates mouse colonization, motility, fimbria-mediated adhesion, and stress tolerance in uropathogenic Proteus mirabilis
Abstract
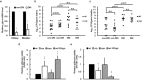
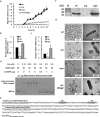
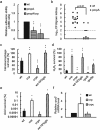
Cyclic AMP receptor protein (Crp) is a major transcriptional regulator in bacteria. This study demonstrated that Crp affects numerous virulence-related phenotypes, including colonization of mice, motility, fimbria-mediated adhesion, and glucose stress tolerance in uropathogenic Proteus mirabilis. Diabetic mice were more susceptible to kidney colonization by wild-type strain than nondiabetic mice, in which the crp mutant exhibited increased kidney colonization. Loss of crp or addition of 10% glucose increased the P. mirabilis adhesion to kidney cells. Direct negative regulation of pmpA (which encodes the major subunit of P-like fimbriae) expression by Crp was demonstrated using a reporter assay and DNase I footprinting. Moreover, the pmpA/crp double mutant exhibited reduced kidney adhesion comparable to that of the pmpA mutant, and mouse kidney colonization by the pmpA mutant was significantly attenuated. Hence, the upregulation of P-like fimbriae in the crp mutant substantially enhanced kidney colonization. Moreover, increased survival in macrophages, increased stress tolerance, RpoS upregulation, and flagellum deficiency leading to immune evasion may promote kidney colonization by the crp mutant. This is the first study to elucidate the role of Crp in the virulence of uropathogenic P. mirabilis, underlying mechanisms, and related therapeutic potential.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
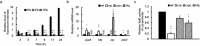

Similar articles
-
Involvement of polyphosphate kinase in virulence and stress tolerance of uropathogenic Proteus mirabilis.Med Microbiol Immunol. 2016 Apr;205(2):97-109. doi: 10.1007/s00430-015-0430-1. Epub 2015 Aug 2. Med Microbiol Immunol. 2016. PMID: 26233310 Free PMC article.
-
Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis.Mol Microbiol. 2002 Aug;45(3):865-74. doi: 10.1046/j.1365-2958.2002.03067.x. Mol Microbiol. 2002. PMID: 12139630
-
Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis.Infect Immun. 1999 Jun;67(6):2822-33. doi: 10.1128/IAI.67.6.2822-2833.1999. Infect Immun. 1999. PMID: 10338487 Free PMC article.
-
Fimbriae of uropathogenic Proteus mirabilis.FEMS Immunol Med Microbiol. 2007 Oct;51(1):1-7. doi: 10.1111/j.1574-695X.2007.00284.x. Epub 2007 Jul 19. FEMS Immunol Med Microbiol. 2007. PMID: 17640292 Review.
-
From Catheter to Kidney Stone: The Uropathogenic Lifestyle of Proteus mirabilis.Trends Microbiol. 2017 Apr;25(4):304-315. doi: 10.1016/j.tim.2016.11.015. Epub 2016 Dec 22. Trends Microbiol. 2017. PMID: 28017513 Free PMC article. Review.
Cited by
-
The QseEF Two-Component System-GlmY Small RNA Regulatory Pathway Controls Swarming in Uropathogenic Proteus mirabilis.Int J Mol Sci. 2022 Jan 1;23(1):487. doi: 10.3390/ijms23010487. Int J Mol Sci. 2022. PMID: 35008912 Free PMC article.
-
Unveiling the hidden arsenal: new insights into Proteus mirabilis virulence in UTIs.Front Cell Infect Microbiol. 2024 Nov 13;14:1465460. doi: 10.3389/fcimb.2024.1465460. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39606746 Free PMC article. Review.
-
Phospholipids and Fatty Acids Affect the Colonization of Urological Catheters by Proteus mirabilis.Int J Mol Sci. 2021 Aug 6;22(16):8452. doi: 10.3390/ijms22168452. Int J Mol Sci. 2021. PMID: 34445157 Free PMC article.
-
cAMP-independent Crp homolog adds to the multi-layer regulatory network in Porphyromonas gingivalis.Front Cell Infect Microbiol. 2025 Apr 16;15:1535009. doi: 10.3389/fcimb.2025.1535009. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40308968 Free PMC article.
-
Virulence genes and antimicrobial resistance pattern in Proteus mirabilis strains isolated from patients attended with urinary infections to Tertiary Hospitals, in Iran.Afr Health Sci. 2021 Dec;21(4):1677-1684. doi: 10.4314/ahs.v21i4.22. Afr Health Sci. 2021. PMID: 35283944 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

