Virome comparisons in wild-diseased and healthy captive giant pandas
- PMID: 28780905
- PMCID: PMC5545856
- DOI: 10.1186/s40168-017-0308-0
Virome comparisons in wild-diseased and healthy captive giant pandas
Abstract
Background: The giant panda (Ailuropoda melanoleuca) is a vulnerable mammal herbivore living wild in central China. Viral infections have become a potential threat to the health of these endangered animals, but limited information related to these infections is available.
Methods: Using a viral metagenomic approach, we surveyed viruses in the feces, nasopharyngeal secretions, blood, and different tissues from a wild giant panda that died from an unknown disease, a healthy wild giant panda, and 46 healthy captive animals.
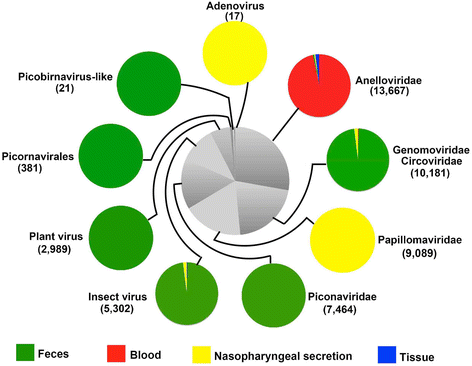
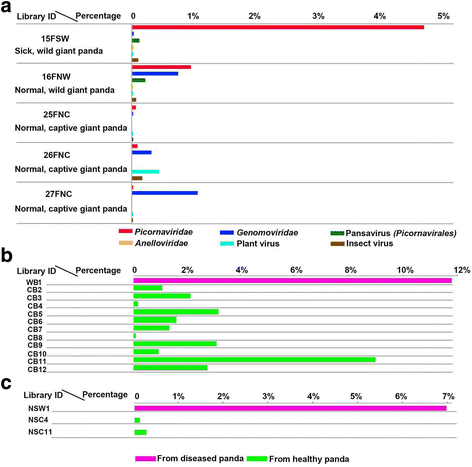
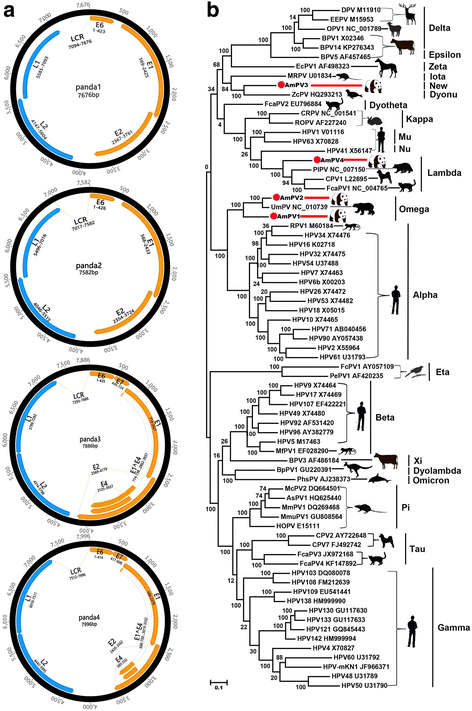
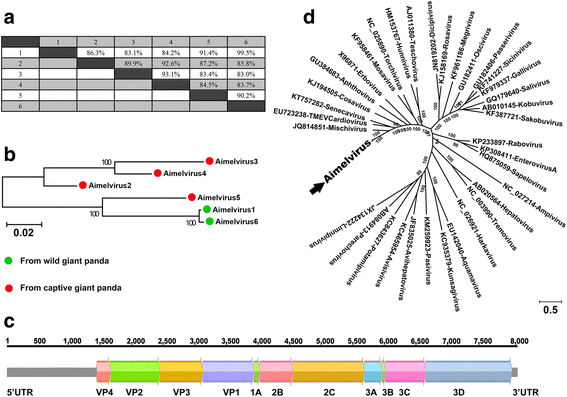
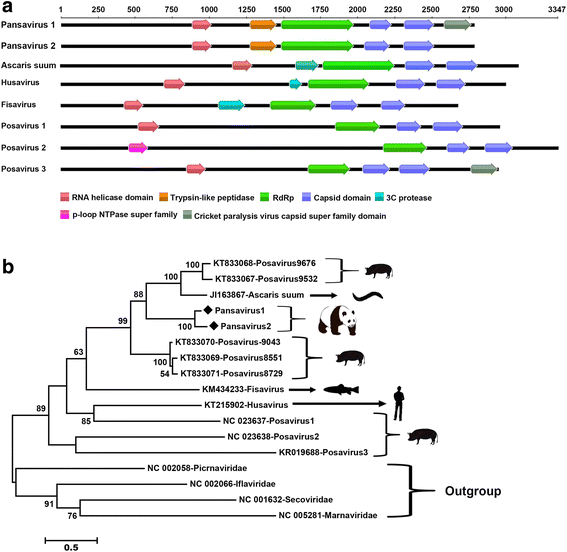
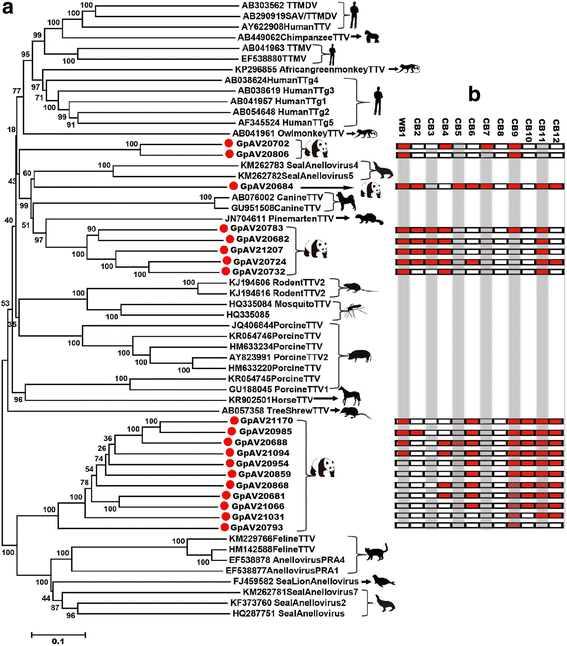
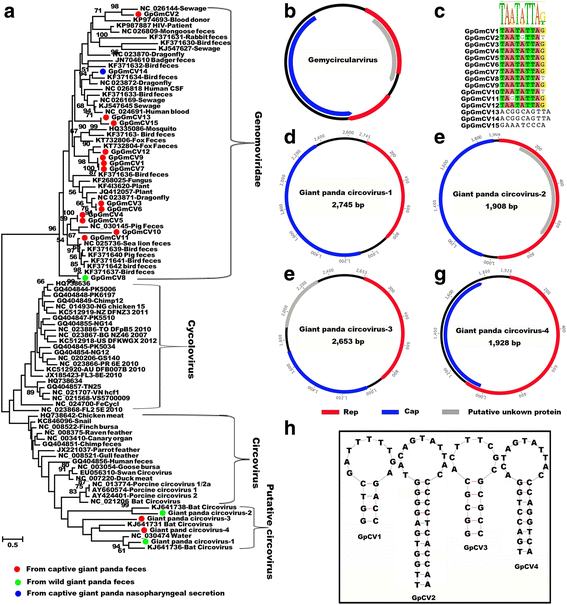
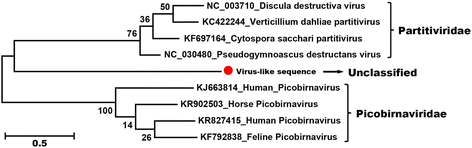
Results: The previously uncharacterized complete or near complete genomes of four viruses from three genera in Papillomaviridae family, six viruses in a proposed new Picornaviridae genus (Aimelvirus), two unclassified viruses related to posaviruses in Picornavirales order, 19 anelloviruses in four different clades of Anelloviridae family, four putative circoviruses, and 15 viruses belonging to the recently described Genomoviridae family were sequenced. Reflecting the diet of giant pandas, numerous insect virus sequences related to the families Iflaviridae, Dicistroviridae, Iridoviridae, Baculoviridae, Polydnaviridae, and subfamily Densovirinae and plant viruses sequences related to the families Tombusviridae, Partitiviridae, Secoviridae, Geminiviridae, Luteoviridae, Virgaviridae, and Rhabdoviridae; genus Umbravirus, Alphaflexiviridae, and Phycodnaviridae were also detected in fecal samples. A small number of insect virus sequences were also detected in the nasopharyngeal secretions of healthy giant pandas and lung tissues from the dead wild giant panda. Although the viral families present in the sick giant panda were also detected in the healthy ones, a higher proportion of papillomaviruses, picornaviruses, and anelloviruses reads were detected in the diseased panda.
Conclusion: This viral survey increases our understanding of eukaryotic viruses in giant pandas and provides a baseline for comparison to viruses detected in future infectious disease outbreaks. The similar viral families detected in sick and healthy giant pandas indicate that these viruses result in commensal infections in most immuno-competent animals.
Keywords: Anellovirus; Gemycircularvirus; Giant panda; Papillomavirus; Picornavirus; Putative circovirus; Viral metagenomics; Virome.
Conflict of interest statement
Ethics approval and consent to participate
The sample collection and all the experiments in the present study were performed with an ethical approval given by Ethics Committee of Jiangsu University and the reference number is No. UJS2014017.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Virome in Fecal Samples From Wild Giant Pandas (Ailuropoda Melanoleuca).Front Vet Sci. 2021 Nov 12;8:767494. doi: 10.3389/fvets.2021.767494. eCollection 2021. Front Vet Sci. 2021. PMID: 34869737 Free PMC article.
-
The fecal viral flora of wild rodents.PLoS Pathog. 2011 Sep;7(9):e1002218. doi: 10.1371/journal.ppat.1002218. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909269 Free PMC article.
-
Virome of Giant Panda-Infesting Ticks Reveals Novel Bunyaviruses and Other Viruses That Are Genetically Close to Those from Giant Pandas.Microbiol Spectr. 2022 Aug 31;10(4):e0203422. doi: 10.1128/spectrum.02034-22. Epub 2022 Aug 2. Microbiol Spectr. 2022. PMID: 35916407 Free PMC article.
-
Review on parasites of wild and captive giant pandas (Ailuropoda melanoleuca): Diversity, disease and conservation impact.Int J Parasitol Parasites Wildl. 2020 Jul 28;13:38-45. doi: 10.1016/j.ijppaw.2020.07.007. eCollection 2020 Dec. Int J Parasitol Parasites Wildl. 2020. PMID: 32793415 Free PMC article. Review.
-
Telemetry research on elusive wildlife: A synthesis of studies on giant pandas.Integr Zool. 2016 Jul;11(4):295-307. doi: 10.1111/1749-4877.12197. Integr Zool. 2016. PMID: 26940046 Review.
Cited by
-
Circovirus in Blood of a Febrile Horse with Hepatitis.Viruses. 2021 May 20;13(5):944. doi: 10.3390/v13050944. Viruses. 2021. PMID: 34065502 Free PMC article.
-
Epidemiological and evolutionary analysis of canine circovirus from 1996 to 2023.BMC Vet Res. 2024 Jul 20;20(1):328. doi: 10.1186/s12917-024-04186-6. BMC Vet Res. 2024. PMID: 39033103 Free PMC article.
-
The virome of the white-winged vampire bat Diaemus youngi is rich in circular DNA viruses.Virus Genes. 2022 Jun;58(3):214-226. doi: 10.1007/s11262-022-01897-6. Epub 2022 Apr 2. Virus Genes. 2022. PMID: 35366197 Free PMC article.
-
Virome of high-altitude canine digestive tract and genetic characterization of novel viruses potentially threatening human health.mSphere. 2023 Oct 24;8(5):e0034523. doi: 10.1128/msphere.00345-23. Epub 2023 Sep 19. mSphere. 2023. PMID: 37724888 Free PMC article.
-
Evolution of anelloviruses from a circovirus-like ancestor through gradual augmentation of the jelly-roll capsid protein.Virus Evol. 2023 May 27;9(1):vead035. doi: 10.1093/ve/vead035. eCollection 2023. Virus Evol. 2023. PMID: 37325085 Free PMC article.
References
-
- Krause J, Unger T, Noçon A, Malaspinas A-S, Kolokotronis S-O, Stiller M, Soibelzon L, Spriggs H, Dear PH, Briggs AW, Bray SCE, O’Brien SJ, Rabeder G, Matheus P, Cooper A, Slatkin M, Pääbo S, Hofreiter M. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

