Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care
- PMID: 28781886
- PMCID: PMC5539963
- DOI: 10.1038/npjgenmed.2016.19
Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care
Abstract
We describe Exome Cancer Test v1.0 (EXaCT-1), the first New York State-Department of Health-approved whole-exome sequencing (WES)-based test for precision cancer care. EXaCT-1 uses HaloPlex (Agilent) target enrichment followed by next-generation sequencing (Illumina) of tumour and matched constitutional control DNA. We present a detailed clinical development and validation pipeline suitable for simultaneous detection of somatic point/indel mutations and copy-number alterations (CNAs). A computational framework for data analysis, reporting and sign-out is also presented. For the validation, we tested EXaCT-1 on 57 tumours covering five distinct clinically relevant mutations. Results demonstrated elevated and uniform coverage compatible with clinical testing as well as complete concordance in variant quality metrics between formalin-fixed paraffin embedded and fresh-frozen tumours. Extensive sensitivity studies identified limits of detection threshold for point/indel mutations and CNAs. Prospective analysis of 337 cancer cases revealed mutations in clinically relevant genes in 82% of tumours, demonstrating that EXaCT-1 is an accurate and sensitive method for identifying actionable mutations, with reasonable costs and time, greatly expanding its utility for advanced cancer care.
Conflict of interest statement
COMPETING INTERESTS The authors declare no conflict of interest.
Figures

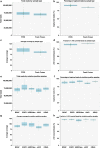
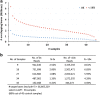

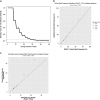

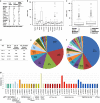
References
-
- Rabbani, B., Tekin, M. & Mahdieh, N. The promise of whole-exome sequencing in medical genetics. J. Hum. Genet. 59, 5–15 (2014). - PubMed
-
- Biesecker, L. G. Exome sequencing makes medical genomics a reality. Nat. Genet. 42, 13–14. - PubMed
-
- Majewski, J., Schwartzentruber, J., Lalonde, E., Montpetit, A. & Jabado, N. What can exome sequencing do for you? J. Med. Genet. 48, 580–589 (2011). - PubMed
-
- Schreiber, M., Dorschner, M. & Tsuang, D. Next-generation sequencing in schizophrenia and other neuropsychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 162B, 671–678 (2013). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

