Deep learning with convolutional neural networks for EEG decoding and visualization
- PMID: 28782865
- PMCID: PMC5655781
- DOI: 10.1002/hbm.23730
Deep learning with convolutional neural networks for EEG decoding and visualization
Abstract
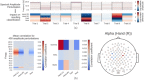
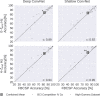
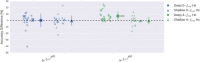
Deep learning with convolutional neural networks (deep ConvNets) has revolutionized computer vision through end-to-end learning, that is, learning from the raw data. There is increasing interest in using deep ConvNets for end-to-end EEG analysis, but a better understanding of how to design and train ConvNets for end-to-end EEG decoding and how to visualize the informative EEG features the ConvNets learn is still needed. Here, we studied deep ConvNets with a range of different architectures, designed for decoding imagined or executed tasks from raw EEG. Our results show that recent advances from the machine learning field, including batch normalization and exponential linear units, together with a cropped training strategy, boosted the deep ConvNets decoding performance, reaching at least as good performance as the widely used filter bank common spatial patterns (FBCSP) algorithm (mean decoding accuracies 82.1% FBCSP, 84.0% deep ConvNets). While FBCSP is designed to use spectral power modulations, the features used by ConvNets are not fixed a priori. Our novel methods for visualizing the learned features demonstrated that ConvNets indeed learned to use spectral power modulations in the alpha, beta, and high gamma frequencies, and proved useful for spatially mapping the learned features by revealing the topography of the causal contributions of features in different frequency bands to the decoding decision. Our study thus shows how to design and train ConvNets to decode task-related information from the raw EEG without handcrafted features and highlights the potential of deep ConvNets combined with advanced visualization techniques for EEG-based brain mapping. Hum Brain Mapp 38:5391-5420, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: EEG analysis; brain mapping; brain-computer interface; brain-machine interface; electroencephalography; end-to-end learning; machine learning; model interpretability.
© 2017 The Authors Human Brain Mapping Published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures










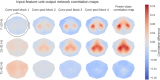
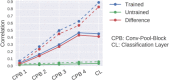


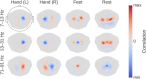
References
-
- Abdel‐Hamid O, Mohamed A. r, Jiang H, Deng L, Penn G, Yu D (2014): Convolutional neural networks for speech recognition. IEEE/ACM Trans Audio Speech Lang Process 22:1533–1545.
-
- Ang KK, Chin ZY, Zhang H, Guan C (2008): Filter Bank Common Spatial Pattern (FBCSP) in Brain‐Computer Interface. In IEEE International Joint Conference on Neural Networks, 2008. IJCNN 2008. (IEEE World Congress on Computational Intelligence), pp 2390–2397.
-
- Antoniades A, Spyrou L, Took CC, Sanei S (2016): Deep learning for epileptic intracranial EEG data. In 2016 IEEE 26th International Workshop on Machine Learning for Signal Processing (MLSP), pp 1–6.
-
- Ball T, Demandt E, Mutschler I, Neitzel E, Mehring C, Vogt K, Aertsen A, Schulze‐Bonhage A (2008): Movement related activity in the high gamma range of the human EEG. NeuroImage 41:302–310. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

